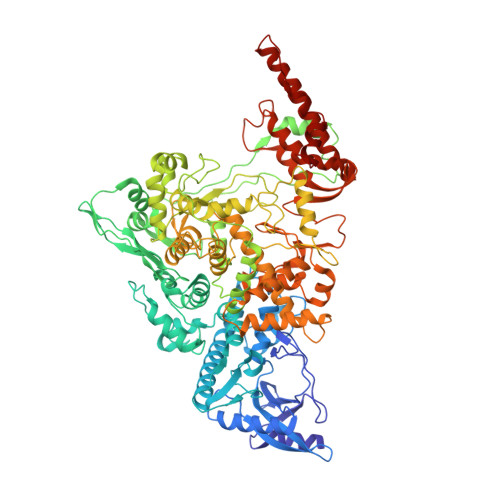
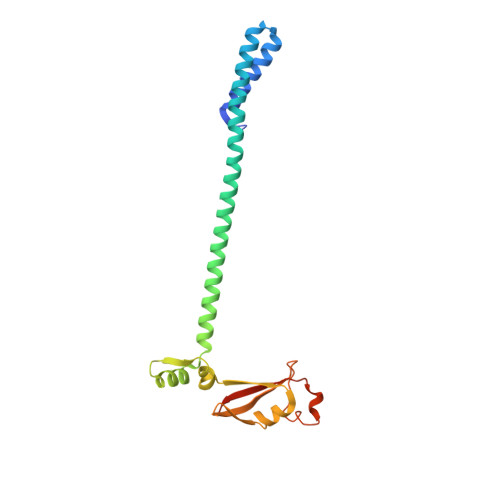
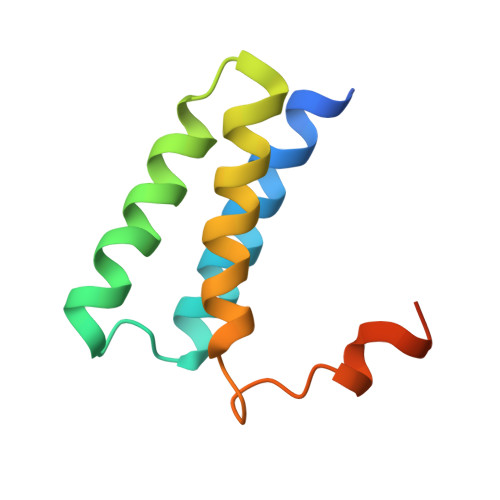
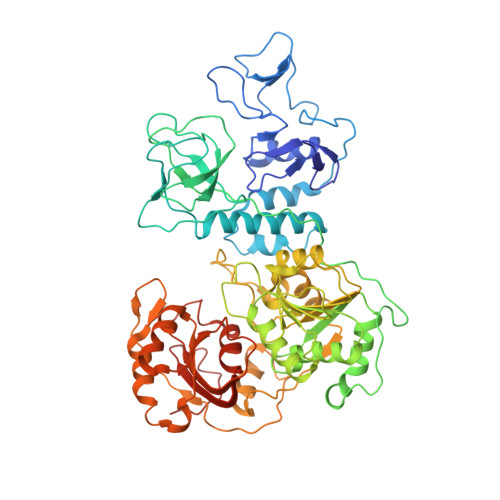
Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Chen, J., Wang, Q., Malone, B., Llewellyn, E., Pechersky, Y., Maruthi, K., Eng, E.T., Perry, J.K., Campbell, E.A., Shaw, D.E., Darst, S.A.(2022) Nat Struct Mol Biol 29: 250-260
- PubMed: 35260847
- DOI: https://doi.org/10.1038/s41594-022-00734-6
- Primary Citation of Related Structures:
7RDX, 7RDY, 7RDZ, 7RE0, 7RE1, 7RE2, 7RE3 - PubMed Abstract:
The SARS-CoV-2 nonstructural proteins coordinate genome replication and gene expression. Structural analyses revealed the basis for coupling of the essential nsp13 helicase with the RNA-dependent RNA polymerase (RdRp) where the holo-RdRp and RNA substrate (the replication-transcription complex or RTC) associated with two copies of nsp13 (nsp13 2 -RTC). One copy of nsp13 interacts with the template-RNA in an opposing polarity to the RdRp and is envisaged to drive the RdRp backward on the RNA template (backtracking), prompting questions as to how the RdRp can efficiently synthesize RNA in the presence of nsp13. Here we use cryogenic-electron microscopy and molecular dynamics simulations to analyze the nsp13 2 -RTC, revealing four distinct conformational states of the helicases. The results indicate a mechanism for the nsp13 2 -RTC to turn backtracking on and off, using an allosteric mechanism to switch between RNA synthesis or backtracking in response to stimuli at the RdRp active site.
Organizational Affiliation:
Laboratory of Molecular Biophysics, The Rockefeller University, New York, NY, USA.