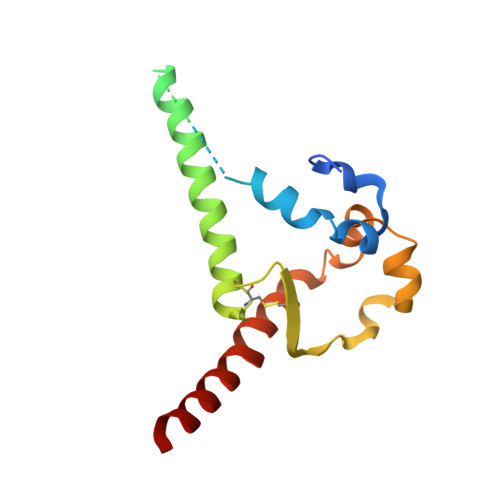
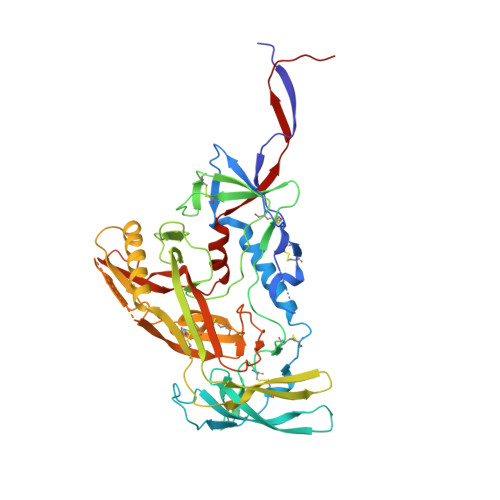
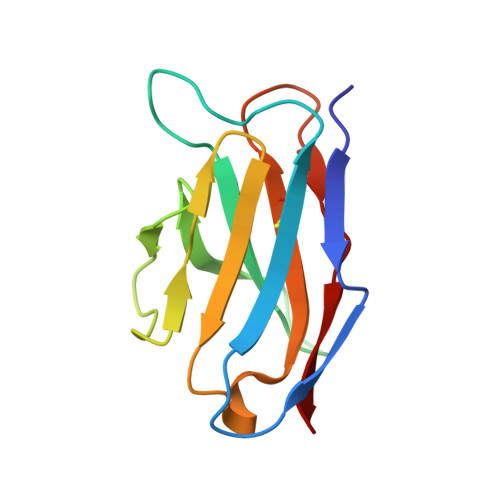
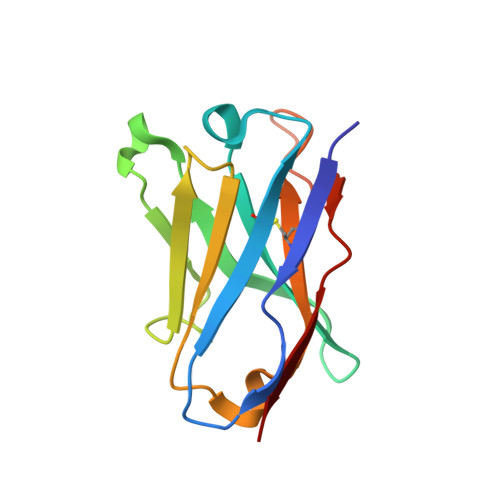
Induction of tier-2 neutralizing antibodies in mice with a DNA-encoded HIV envelope native like trimer.
Xu, Z., Walker, S., Wise, M.C., Chokkalingam, N., Purwar, M., Moore, A., Tello-Ruiz, E., Wu, Y., Majumdar, S., Konrath, K.M., Kulkarni, A., Tursi, N.J., Zaidi, F.I., Reuschel, E.L., Patel, I., Obeirne, A., Du, J., Schultheis, K., Gites, L., Smith, T., Mendoza, J., Broderick, K.E., Humeau, L., Pallesen, J., Weiner, D.B., Kulp, D.W.(2022) Nat Commun 13: 695-695
- PubMed: 35121758
- DOI: https://doi.org/10.1038/s41467-022-28363-z
- Primary Citation of Related Structures:
7SQ1 - PubMed Abstract:
HIV Envelope (Env) is the main vaccine target for induction of neutralizing antibodies. Stabilizing Env into native-like trimer (NLT) conformations is required for recombinant protein immunogens to induce autologous neutralizing antibodies(nAbs) against difficult to neutralize HIV strains (tier-2) in rabbits and non-human primates. Immunizations of mice with NLTs have generally failed to induce tier-2 nAbs. Here, we show that DNA-encoded NLTs fold properly in vivo and induce autologous tier-2 nAbs in mice. DNA-encoded NLTs also uniquely induce both CD4 + and CD8 + T-cell responses as compared to corresponding protein immunizations. Murine neutralizing antibodies are identified with an advanced sequencing technology. The structure of an Env-Ab (C05) complex, as determined by cryo-EM, identifies a previously undescribed neutralizing Env C3/V5 epitope. Beyond potential functional immunity gains, DNA vaccines permit in vivo folding of structured antigens and provide significant cost and speed advantages for enabling rapid evaluation of new HIV vaccines.
Organizational Affiliation:
Vaccine and Immunotherapy Center, The Wistar Institute, Philadelphia, PA, 19104, USA.