Receptor-specific recognition of NPY peptides revealed by structures of NPY receptors.
Tang, T., Tan, Q., Han, S., Diemar, A., Lobner, K., Wang, H., Schuss, C., Behr, V., Morl, K., Wang, M., Chu, X., Yi, C., Keller, M., Kofoed, J., Reedtz-Runge, S., Kaiser, A., Beck-Sickinger, A.G., Zhao, Q., Wu, B.(2022) Sci Adv 8: eabm1232-eabm1232
- PubMed: 35507650
- DOI: https://doi.org/10.1126/sciadv.abm1232
- Primary Citation of Related Structures:
7X9A, 7X9B, 7X9C - PubMed Abstract:
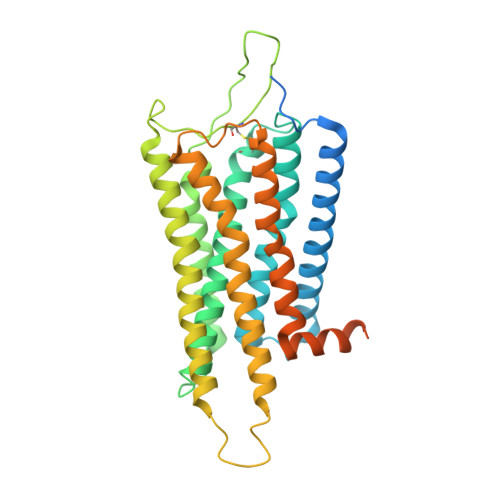
In response to three highly conserved neuropeptides, neuropeptide Y (NPY), peptide YY, and pancreatic polypeptide (PP), four G protein-coupled receptors mediate multiple essential physiological processes, such as food intake, vasoconstriction, sedation, and memory retention. Here, we report the structures of the human Y 1 , Y 2 , and Y 4 receptors in complex with NPY or PP, and the G i1 protein. These structures reveal distinct binding poses of the peptide upon coupling to different receptors, reflecting the importance of the conformational plasticity of the peptide in recognizing the NPY receptors. The N terminus of the peptide forms extensive interactions with the Y 1 receptor, but not with the Y 2 and Y 4 receptors. Supported by mutagenesis and functional studies, subtype-specific interactions between the receptors and peptides were further observed. These findings provide insight into key factors that govern NPY signal recognition and transduction, and would enable development of selective drugs.
Organizational Affiliation:
School of Pharmaceutical Science and Technology, Hangzhou Institute for Advanced Study, UCAS, Hangzhou, China.