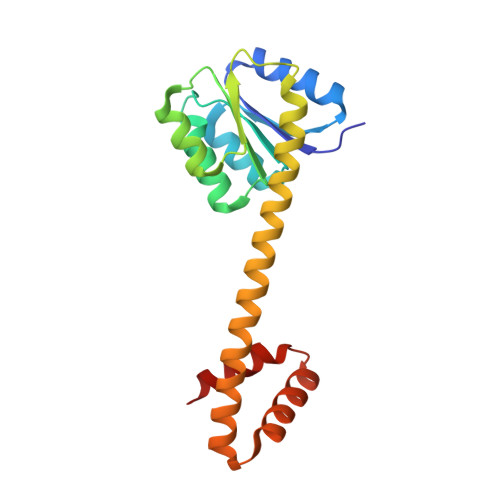
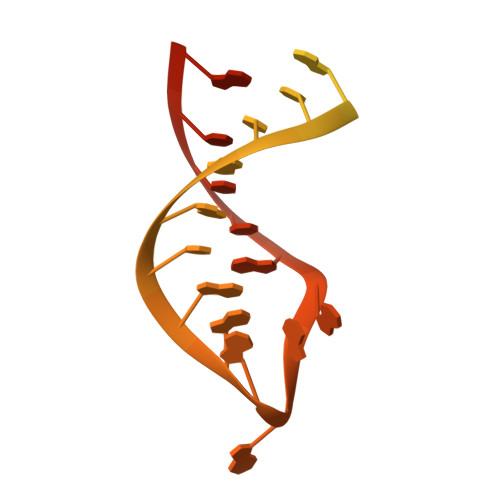
Structural characterization of the ANTAR antiterminator domain bound to RNA.
Walshe, J.L., Siddiquee, R., Patel, K., Ataide, S.F.(2022) Nucleic Acids Res 50: 2889-2904
- PubMed: 35150565
- DOI: https://doi.org/10.1093/nar/gkac074
- Primary Citation of Related Structures:
6WSH, 6WW6 - PubMed Abstract:
Regulated transcription termination provides an efficient and responsive means to control gene expression. In bacteria, rho-independent termination occurs through the formation of an intrinsic RNA terminator loop, which disrupts the RNA polymerase elongation complex, resulting in its dissociation from the DNA template. Bacteria have a number of pathways for overriding termination, one of which is the formation of mutually exclusive RNA motifs. ANTAR domains are a class of antiterminator that bind and stabilize dual hexaloop RNA motifs within the nascent RNA chain to prevent terminator loop formation. We have determined the structures of the dimeric ANTAR domain protein EutV, from Enterococcus faecialis, in the absence of and in complex with the dual hexaloop RNA target. The structures illustrate conformational changes that occur upon RNA binding and reveal that the molecular interactions between the ANTAR domains and RNA are restricted to a single hexaloop of the motif. An ANTAR domain dimer must contact each hexaloop of the dual hexaloop motif individually to prevent termination in eubacteria. Our findings thereby redefine the minimal ANTAR domain binding motif to a single hexaloop and revise the current model for ANTAR-mediated antitermination. These insights will inform and facilitate the discovery of novel ANTAR domain RNA targets.
Organizational Affiliation:
School of Life and Environmental Science, Faculty of Science, University of Sydney, Sydney, NSW 2006, Australia.