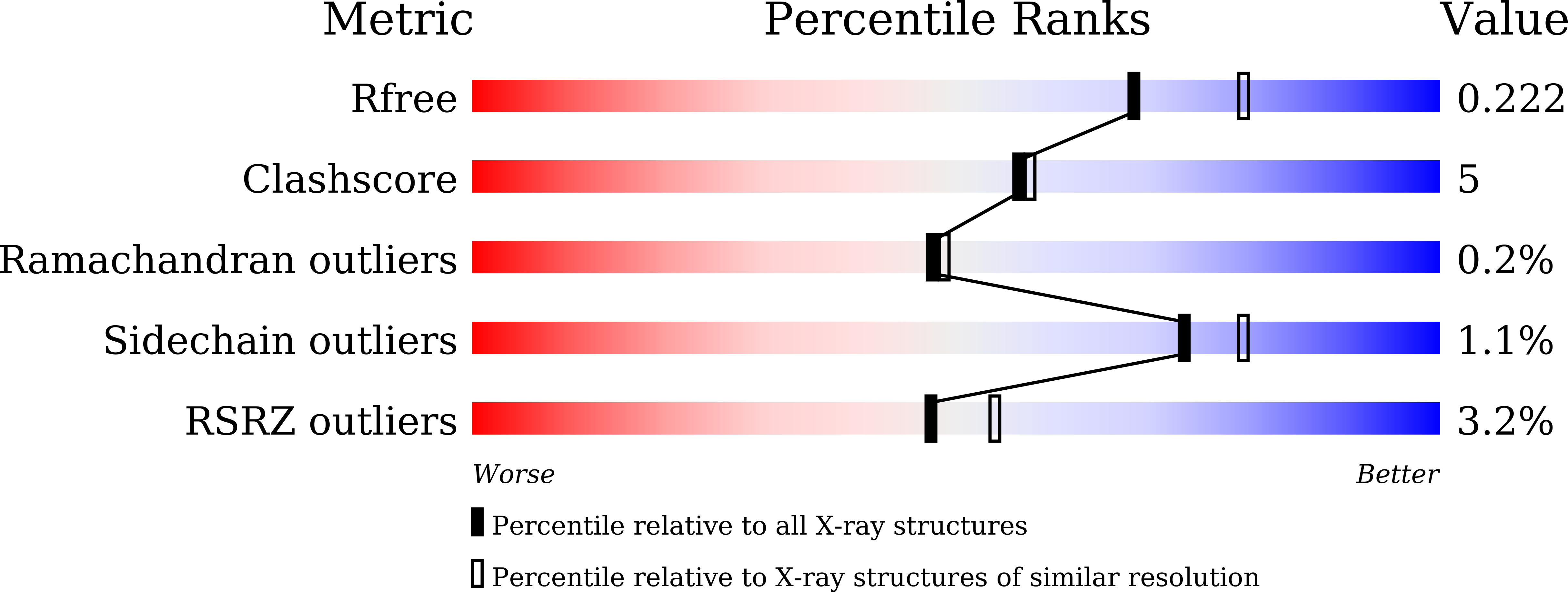
Substantial Improvement of an Epimerase for the Synthesis of D-Allulose by Biosensor-Based High-Throughput Microdroplet Screening
Li, C., Gao, X., Qi, H., Zhang, W., Li, L., Wei, C., Wei, M., Sun, X., Wang, S., Wang, L., Ji, Y., Mao, S., Zhu, Z., Tanokura, M., Lu, F., Qin, H.M.(2023) Angew Chem Int Ed Engl