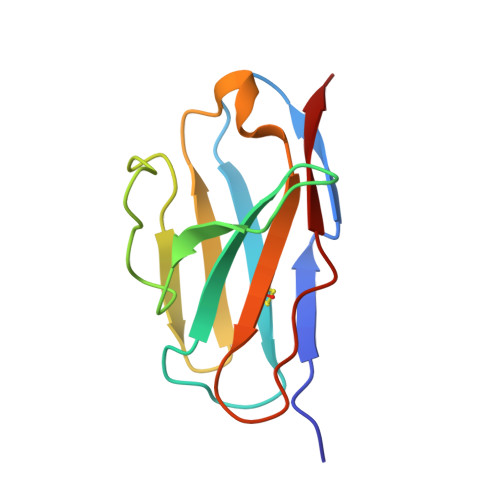
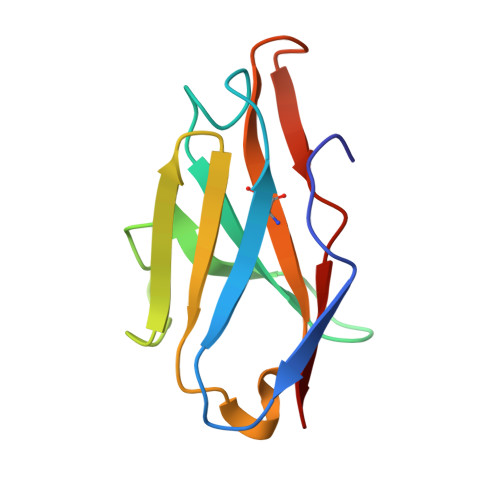
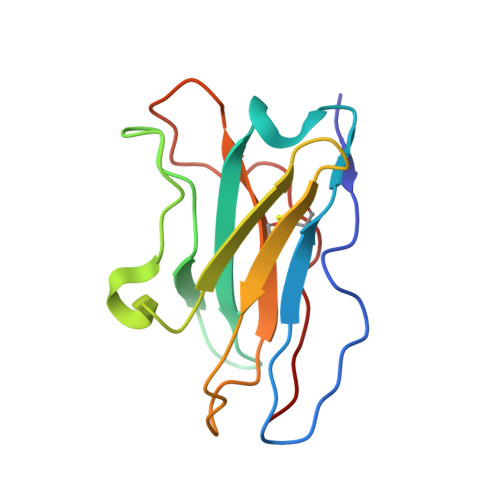
Structure-based development of three- and four-antibody cocktails against SARS-CoV-2 via multiple mechanisms.
Sun, Y., Wang, L., Feng, R., Wang, N., Wang, Y., Zhu, D., Xing, X., Yang, P., Zhang, Y., Li, W., Wang, X.(2021) Cell Res 31: 597-600
- PubMed: 33782529
- DOI: https://doi.org/10.1038/s41422-021-00497-7
- Primary Citation of Related Structures:
7E5R, 7E5S
Organizational Affiliation:
CAS Key Laboratory of Infection and Immunity, National Laboratory of Macromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, 100101, China.