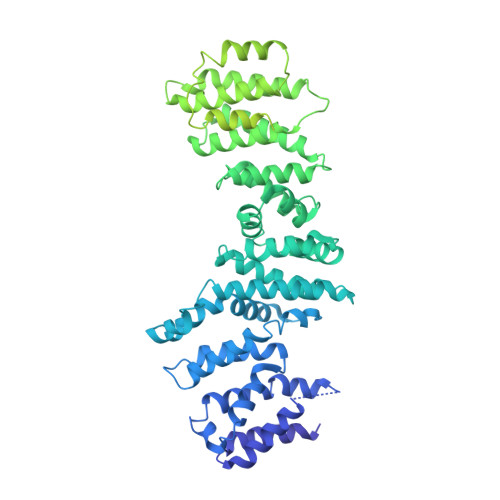
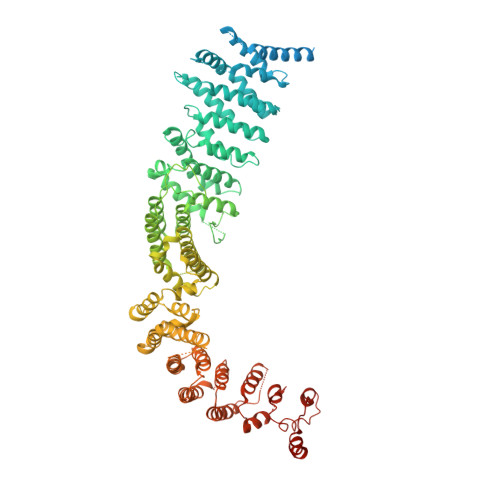
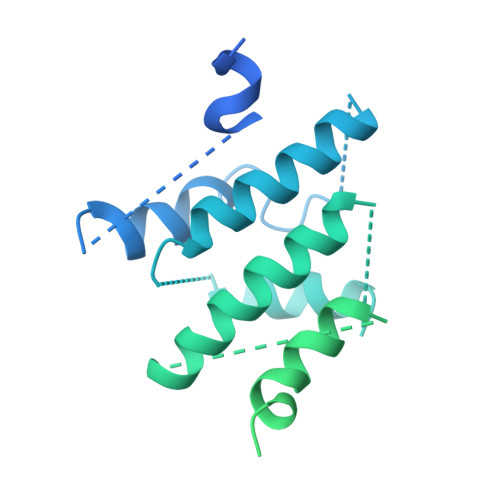
Structure of the Human TELO2-TTI1-TTI2 Complex.
Kim, Y., Park, J., Joo, S.Y., Kim, B.G., Jo, A., Lee, H., Cho, Y.(2022) J Mol Biol 434: 167370-167370
- PubMed: 34838521
- DOI: https://doi.org/10.1016/j.jmb.2021.167370
- Primary Citation of Related Structures:
7F4U - PubMed Abstract:
Phosphatidylinositol 3-kinase-related protein kinases (PIKKs) play critical roles in various metabolic pathways related to cell proliferation and survival. The TELO2-TTI1-TTI2 (TTT) complex has been proposed to recognize newly synthesized PIKKs and to deliver them to the R2TP complex (RUVBL1-RUVBL2-RPAP3-PIH1D1) and the heat shock protein 90 chaperone, thereby supporting their folding and assembly. Here, we determined the cryo-EM structure of the TTT complex at an average resolution of 4.2 Å. We describe the full-length structures of TTI1 and TELO2, and a partial structure of TTI2. All three proteins form elongated helical repeat structures. TTI1 provides a platform on which TELO2 and TTI2 bind to its central region and C-terminal end, respectively. The TELO2 C-terminal domain (CTD) is required for the interaction with TTI1 and recruitment of Ataxia-telangiectasia mutated (ATM). The N- and C-terminal segments of TTI1 recognize the FRAP-ATM-TRRAP (FAT) domain and the N-terminal HEAT repeats of ATM, respectively. The TELO2 CTD and TTI1 N- and C-terminal segments are required for cell survival in response to ionizing radiation.
Organizational Affiliation:
Department of Life Science, Pohang University of Science and Technology, Pohang 37673, South Korea.