Structural basis of sodium-dependent bile salt uptake into the liver.
Goutam, K., Ielasi, F.S., Pardon, E., Steyaert, J., Reyes, N.(2022) Nature 606: 1015-1020
- PubMed: 35545671
- DOI: https://doi.org/10.1038/s41586-022-04723-z
- Primary Citation of Related Structures:
7PQG, 7PQQ - PubMed Abstract:
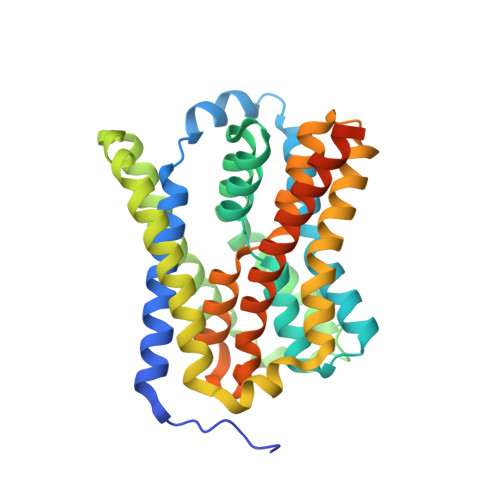
The liver takes up bile salts from blood to generate bile, enabling absorption of lipophilic nutrients and excretion of metabolites and drugs 1 . Human Na + -taurocholate co-transporting polypeptide (NTCP) is the main bile salt uptake system in liver. NTCP is also the cellular entry receptor of human hepatitis B and D viruses 2,3 (HBV/HDV), and has emerged as an important target for antiviral drugs 4 . However, the molecular mechanisms underlying NTCP transport and viral receptor functions remain incompletely understood. Here we present cryo-electron microscopy structures of human NTCP in complexes with nanobodies, revealing key conformations of its transport cycle. NTCP undergoes a conformational transition opening a wide transmembrane pore that serves as the transport pathway for bile salts, and exposes key determinant residues for HBV/HDV binding to the outside of the cell. A nanobody that stabilizes pore closure and inward-facing states impairs recognition of the HBV/HDV receptor-binding domain preS1, demonstrating binding selectivity of the viruses for open-to-outside over inward-facing conformations of the NTCP transport cycle. These results provide molecular insights into NTCP 'gated-pore' transport and HBV/HDV receptor recognition mechanisms, and are expected to help with development of liver disease therapies targeting NTCP.
Organizational Affiliation:
Membrane Protein Mechanisms Group, European Institute of Chemistry and Biology, University of Bordeaux, CNRS-UMR5234, Pessac, France.