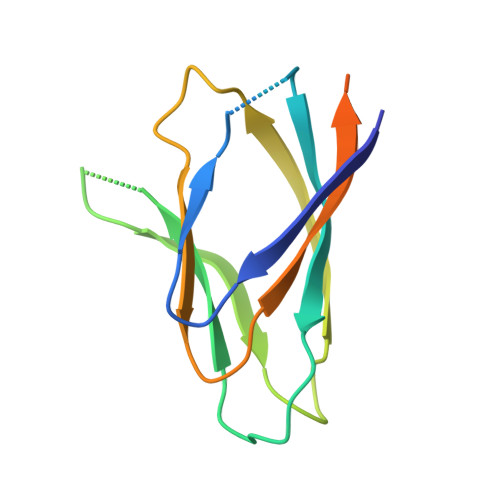
The plant nucleoplasmin AtFKBP43 needs its extended arms for histone interaction.
Singh, A.K., Saharan, K., Baral, S., Vasudevan, D.(2022) Biochim Biophys Acta Gene Regul Mech 1865: 194872-194872
- PubMed: 36058470
- DOI: https://doi.org/10.1016/j.bbagrm.2022.194872
- Primary Citation of Related Structures:
7WIM - PubMed Abstract:
The nucleoplasmin family of histone chaperones is a key player in governing the dynamic architecture of chromatin, thereby regulating various DNA-templated processes. The crystal structure of the N-terminal domain of Arabidopsis thaliana FKBP43 (AtFKBP43), an FK506-binding immunophilin protein, revealed a characteristic nucleoplasmin fold, thus confirming it to be a member of the FKBP nucleoplasmin class. Small-Angle X-ray Scattering (SAXS) analyses confirmed its pentameric nature in solution, and additional studies confirmed the nucleoplasmin fold to be highly stable. Unlike its homolog AtFKBP53, the AtFKBP43 nucleoplasmin core domain could not interact with histones and required the acidic arms, C-terminal to the core, for histone association. However, SAXS generated low-resolution envelope structure, ITC, and AUC results revealed that an AtFKBP43 pentamer with C-terminal extensions interacts with H2A/H2B dimer and H3/H4 tetramer in an equimolar ratio, like AtFKBP53. Put together, AtFKBP43 belongs to a hitherto unreported subclass of FKBP nucleoplasmins that requires the C-terminal acidic stretches emanating from the core domain for histone interaction.
Organizational Affiliation:
Institute of Life Sciences, Bhubaneswar 751023, India.