In situ structures of polymerase complex of mammalian reovirus illuminate RdRp activation and transcription regulation.
Bao, K., Zhang, X., Li, D., Sun, W., Sun, Z., Wang, J., Zhu, P.(2022) Proc Natl Acad Sci U S A 119: e2203054119-e2203054119
- PubMed: 36469786
- DOI: https://doi.org/10.1073/pnas.2203054119
- Primary Citation of Related Structures:
7YED, 7YEV, 7YEZ, 7YF0, 7YFE - PubMed Abstract:
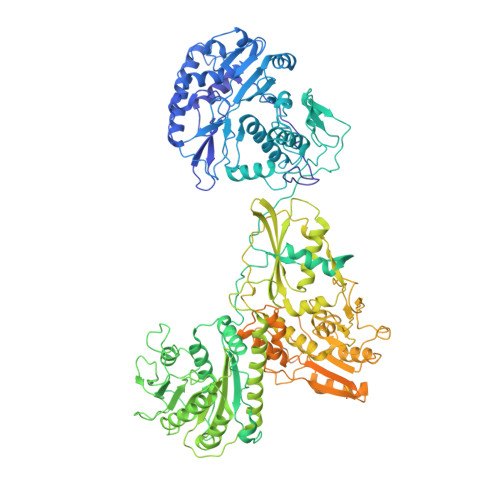
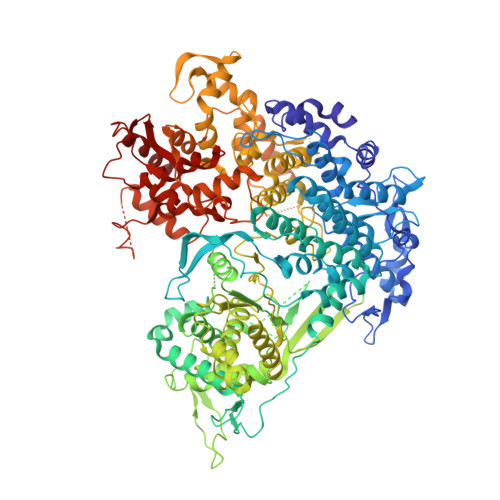
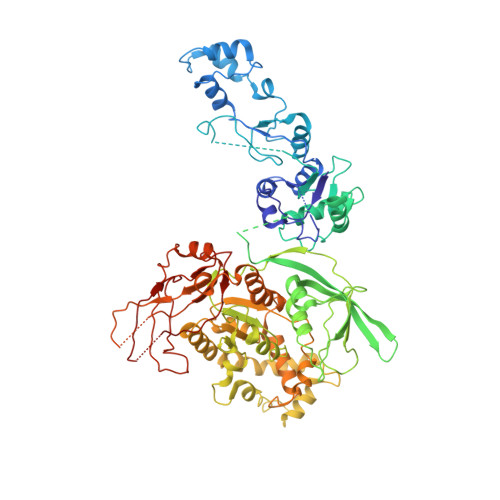
Mammalian reovirus (reovirus) is a multilayered, turreted member of Reoviridae characterized by transcription of dsRNA genome within the innermost capsid shell. Here, we present high-resolution in situ structures of reovirus transcriptase complex in an intact double-layered virion, and in the uncoated single-layered core particles in the unloaded, reloaded, pre-elongation, and elongation states, respectively, obtained by cryo-electron microscopy and sub-particle reconstructions. At the template entry of RNA-dependent RNA polymerase (RdRp), the RNA-loading region gets flexible after uncoating resulting in the unloading of terminal genomic RNA and inactivity of transcription. However, upon adding transcriptional substrates, the RNA-loading region is recovered leading the RNAs loaded again. The priming loop in RdRp was found to play a critical role in regulating transcription, which hinders the elongation of transcript in virion and triggers the rearrangement of RdRp C-terminal domain (CTD) during elongation, resulting in splitting of template-transcript hybrid and opening of transcript exit. With the integration of these structures, a transcriptional model of reovirus with five states is proposed. Our structures illuminate the RdRp activation and regulation of the multilayered turreted reovirus.
Organizational Affiliation:
National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.