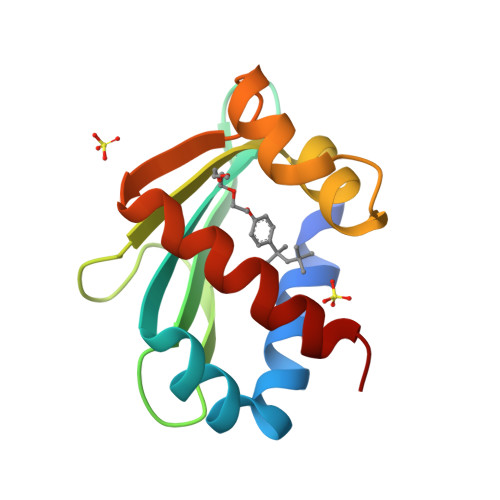
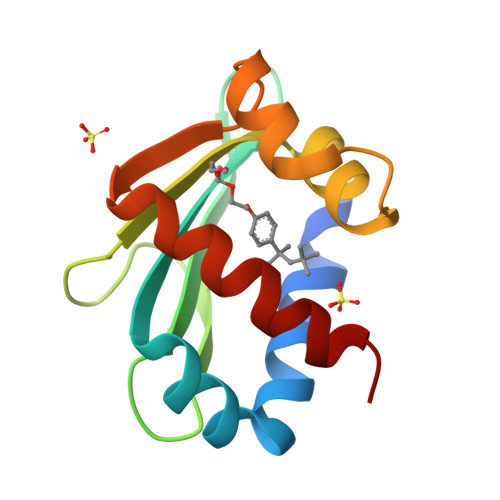
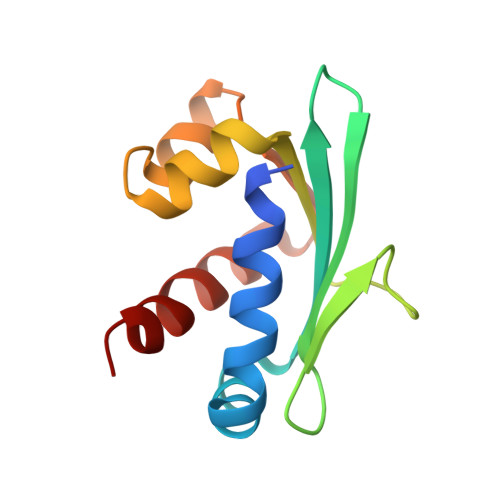
Crystal structure of the liganded SCP-2-like domain of human peroxisomal multifunctional enzyme type 2 at 1.75 A resolution.
Haapalainen, A.M., van Aalten, D.M., Merilainen, G., Jalonen, J.E., Pirila, P., Wierenga, R.K., Hiltunen, J.K., Glumoff, T.(2001) J Mol Biology 313: 1127-1138
- PubMed: 11700068
- DOI: https://doi.org/10.1006/jmbi.2001.5084
- Primary Citation of Related Structures:
1IKT - PubMed Abstract:
beta-Oxidation of amino acyl coenzyme A (acyl-CoA) species in mammalian peroxisomes can occur via either multifunctional enzyme type 1 (MFE-1) or type 2 (MFE-2), both of which catalyze the hydration of trans-2-enoyl-CoA and the dehydrogenation of 3-hydroxyacyl-CoA, but with opposite chiral specificity. MFE-2 has a modular organization of three domains. The function of the C-terminal domain of the mammalian MFE-2, which shows similarity with sterol carrier protein type 2 (SCP-2), is unclear. Here, the structure of the SCP-2-like domain comprising amino acid residues 618-736 of human MFE-2 (d Delta h Delta SCP-2L) was solved at 1.75 A resolution in complex with Triton X-100, an analog of a lipid molecule. This is the first reported structure of an MFE-2 domain. The d Delta h Delta SCP-2L has an alpha/beta-fold consisting of five beta-strands and five alpha-helices; the overall architecture resembles the rabbit and human SCP-2 structures. However, the structure of d Delta h Delta SCP-2L shows a hydrophobic tunnel that traverses the protein, which is occupied by an ordered Triton X-100 molecule. The tunnel is large enough to accommodate molecules such as straight-chain and branched-chain fatty acyl-CoAs and bile acid intermediates. Large empty apolar cavities are observed near the exit of the tunnel and between the helices C and D. In addition, the C-terminal peroxisomal targeting signal is ordered in the structure and solvent-exposed, which is not the case with unliganded rabbit SCP-2, supporting the hypothesis of a ligand-assisted targeting mechanism.
Organizational Affiliation:
Biocenter Oulu and Department of Biochemistry, University of Oulu, FIN-90014, Finland.