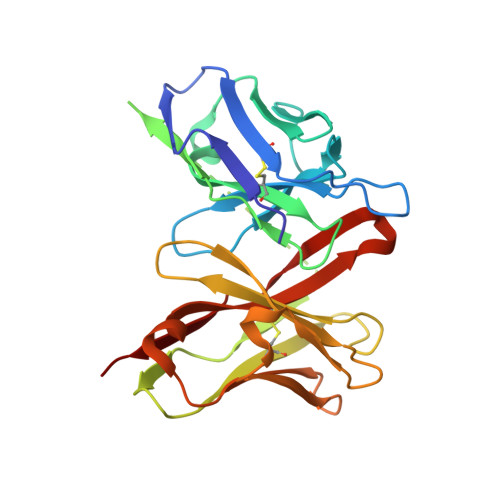
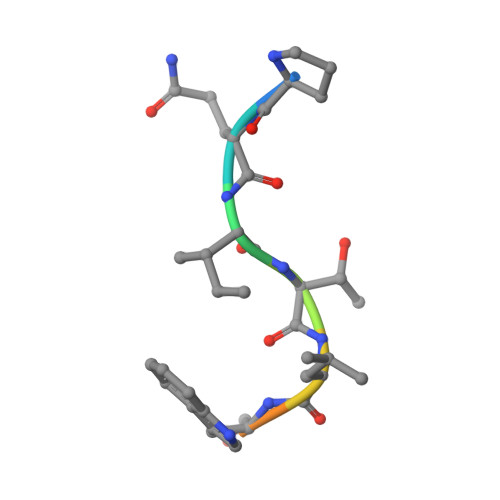
Structural basis of HIV-1 and HIV-2 protease inhibition by a monoclonal antibody.
Rezacova, P., Lescar, J., Brynda, J., Fabry, M., Horejsi, M., Sedlacek, J., Bentley, G.A.(2001) Structure 9: 887-895
- PubMed: 11591344
- DOI: https://doi.org/10.1016/s0969-2126(01)00654-2
- Primary Citation of Related Structures:
1JP5 - PubMed Abstract:
Since the demonstration that the protease of the human immunodeficiency virus (HIV Pr) is essential in the viral life cycle, this enzyme has become one of the primary targets for antiviral drug design. The murine monoclonal antibody 1696 (mAb1696), produced by immunization with the HIV-1 protease, inhibits the catalytic activity of the enzyme of both the HIV-1 and HIV-2 isolates with inhibition constants in the low nanomolar range. The antibody cross-reacts with peptides that include the N terminus of the enzyme, a region that is highly conserved in sequence among different viral strains and that, furthermore, is crucial for homodimerization to the active enzymatic form. We report here the crystal structure at 2.7 A resolution of a recombinant single-chain Fv fragment of mAb1696 as a complex with a cross-reactive peptide of the HIV-1 protease. The antibody-antigen interactions observed in this complex provide a structural basis for understanding the origin of the broad reactivity of mAb-1696 for the HIV-1 and HIV-2 proteases and their respective N-terminal peptides. A possible mechanism of HIV-protease inhibition by mAb1696 is proposed that could help the design of inhibitors aimed at binding inactive monomeric species.
Organizational Affiliation:
Department of Gene Manipulation, Institute of Molecular Genetics, Academy of Sciences of the Czech Republic, Flemingovo nam. 2, 166 37 Prague 6, Czech Republic. rezacova@img.cas.cz