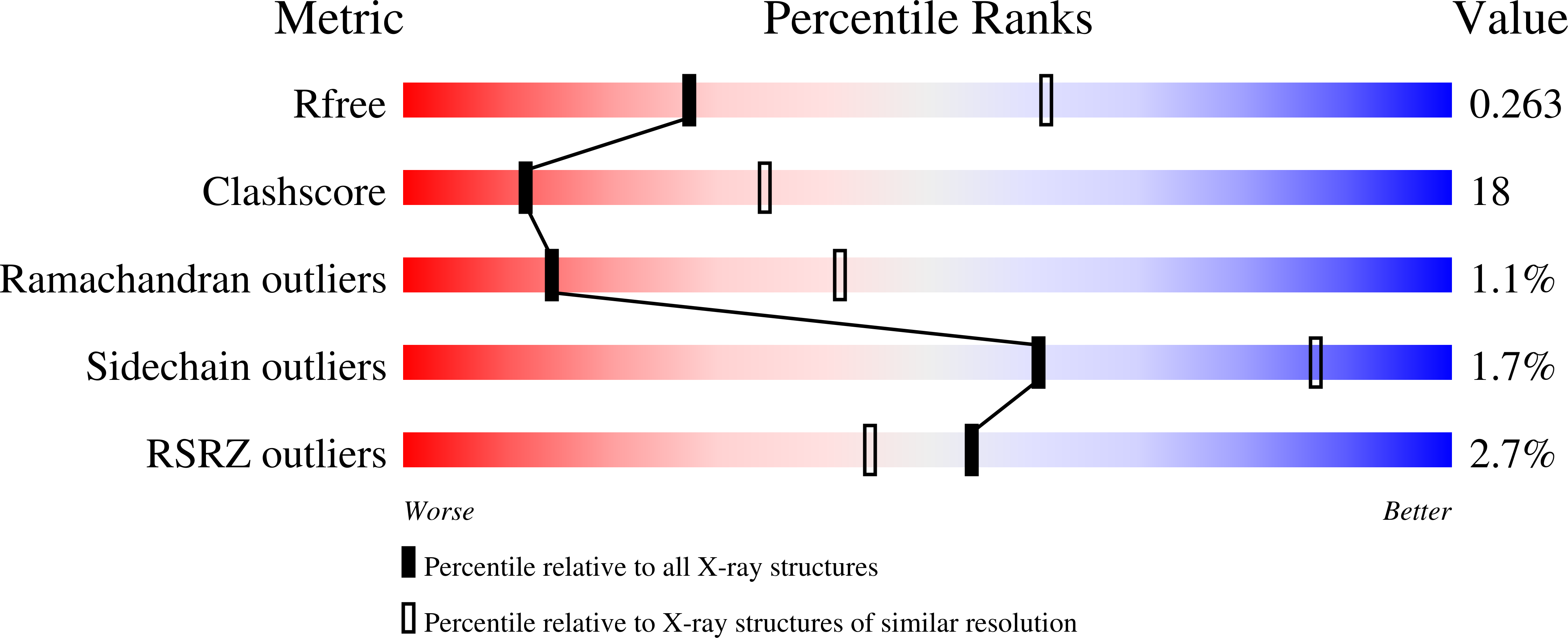
Novel class III phosphoribosyl diphosphate synthase: structure and properties of the tetrameric, phosphate-activated, non-allosterically inhibited enzyme from Methanocaldococcus jannaschii
Kadziola, A., Jepsen, C.H., Johansson, E., McGuire, J., Larsen, S., Hove-Jensen, B.(2005) J Mol Biol 354: 815-828
- PubMed: 16288921
- DOI: https://doi.org/10.1016/j.jmb.2005.10.001
- Primary Citation of Related Structures:
1U9Y, 1U9Z - PubMed Abstract:
The prs gene encoding phosphoribosyl diphosphate (PRPP) synthase of the hyperthermophilic autotrophic methanogenic archaeon Methanocaldococcus jannaschii has been cloned and expressed in Escherichia coli. Subsequently, M.jannaschii PRPP synthase has been purified, characterised, crystallised, and the crystal structure determined. The enzyme is activated by phosphate ions and only ATP or dATP serve as diphosphoryl donors. The K(m) values are determined as 2.6 mM and 2.8 mM for ATP and ribose 5-phosphate, respectively, and the V(max) value as 2.20 mmol (minxmg of protein)(-1). ADP is a potent inhibitor of activity while GDP has no effect. A single ADP binding site, the active site, is present per subunit. The crystal structure of the enzyme reveals a more compact subunit than that of the enzyme from the mesophile Bacillus subtilis, caused by truncations at the N and C terminus as well as shorter loops in the M.jannaschii enzyme. The M.jannaschii enzyme displays a tetrameric quaternary structure in contrast to the hexameric quaternary structure of B.subtilis PRPP synthase. Soaking of the crystals with 5'-AMP and PRPP revealed the position of the former compound as well as that of ribose 5-phosphate. The properties of M.jannaschii PRPP synthase differ widely from previously characterised PRPP synthases by its tetrameric quaternary structure and the simultaneous phosphate ion-activation and lack of allosteric inhibition, and, thus, constitute a novel class of PRPP synthases.
Organizational Affiliation:
Centre for Crystallographic Studies, Department of Chemistry, University of Copenhagen, 5 Universitetsparken, DK-2100 Copenhagen Ø, Denmark. anders@ccs.ki.ku.dk