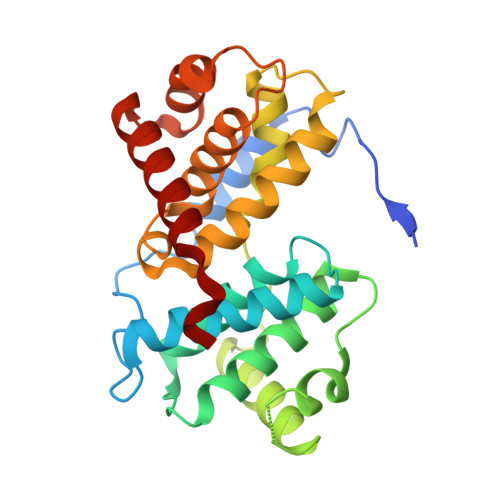
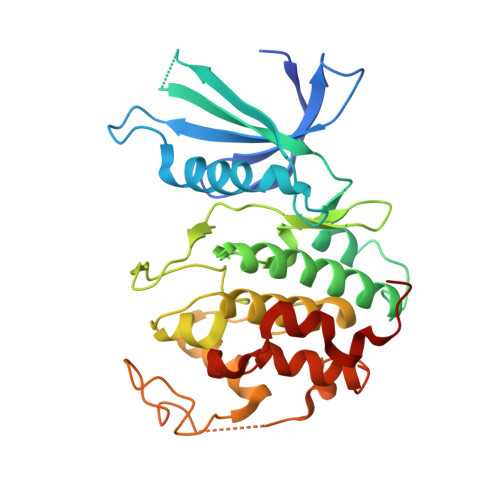
Crystal Structure of a Human Cyclin-Dependent Kinase 6 Complex with a Flavonol Inhibitor, Fisetin.
Lu, H.S., Chang, D.J., Baratte, B., Meijer, L., Schulze-Gahmen, U.(2005) J Med Chem 48: 737-747
- PubMed: 15689157
- DOI: https://doi.org/10.1021/jm049353p
- Primary Citation of Related Structures:
1XO2 - PubMed Abstract:
Cyclin-dependent kinases (CDKs) play a central role in cell cycle control, apoptosis, transcription, and neuronal functions. They are important targets for the design of drugs with antimitotic or antineurodegenerative effects. CDK4 and CDK6 form a subfamily among the CDKs in mammalian cells, as defined by sequence similarities. Compared to CDK2 and CDK5, structural information on CDK4 and CDK6 is sparse. We describe here the crystal structure of human CDK6 in complex with a viral cyclin and a flavonol inhibitor, fisetin. Fisetin binds to the active form of CDK6, forming hydrogen bonds with the side chains of residues in the binding pocket that undergo large conformational changes during CDK activation by cyclin binding. The 4-keto group and the 3-hydroxyl group of fisetin are hydrogen bonded with the backbone in the hinge region between the N-terminal and C-terminal kinase domain, as has been observed for many CDK inhibitors. However, CDK2 and HCK kinase in complex with other flavone inhibitors such as quercetin and flavopiridol showed a different binding mode with the inhibitor rotated by about 180 degrees. The structural information of the CDK6-fisetin complex is correlated with the binding affinities of different flavone inhibitors for CDK6. This complex structure is the first description of an inhibitor complex with a kinase from the CDK4/6 subfamily and can provide a basis for selecting and designing inhibitor compounds with higher affinities and specificities.
- Physical Biosciences Division, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, MS 64R0121, Berkeley, California 94720, USA.
Organizational Affiliation: