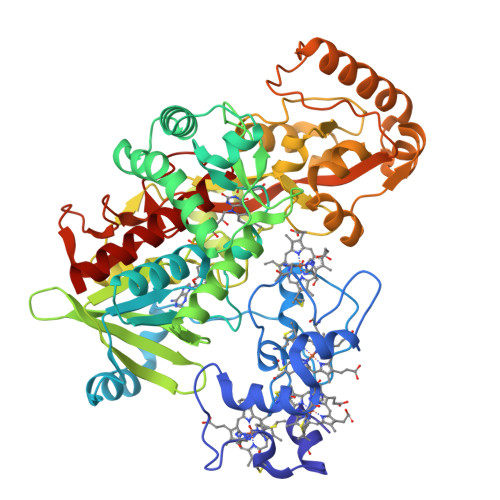
A Proton Delivery Pathway in the Soluble Fumarate Reductase from Shewanella frigidimarina.
Pankhurst, K.L., Mowat, C.G., Rothery, E.L., Hudson, J.M., Jones, A.K., Miles, C.S., Walkinshaw, M.D., Armstrong, F.A., Reid, G.A., Chapman, S.K.(2006) J Biological Chem 281: 20589-20597
- PubMed: 16699170
- DOI: https://doi.org/10.1074/jbc.M603077200
- Primary Citation of Related Structures:
2B7R, 2B7S - PubMed Abstract:
The mechanism for fumarate reduction by the soluble fumarate reductase from Shewanella frigidimarina involves hydride transfer from FAD and proton transfer from the active-site acid, Arg-402. It has been proposed that Arg-402 forms part of a proton transfer pathway that also involves Glu-378 and Arg-381 but, unusually, does not involve any bound water molecules. To gain further insight into the importance of this proton pathway we have perturbed it by substituting Arg-381 by lysine and methionine and Glu-378 by aspartate. Although all the mutant enzymes retain measurable activities, there are orders-of-magnitude decreases in their k(cat) values compared with the wild-type enzyme. Solvent kinetic isotope effects show that proton transfer is rate-limiting in the wild-type and mutant enzymes. Proton inventories indicate that the proton pathway involves multiple exchangeable groups. Fast scan protein-film voltammetric studies on wild-type and R381K enzymes show that the proton transfer pathway delivers one proton per catalytic cycle and is not required for transporting the other proton, which transfers as a hydride from the reduced, protonated FAD. The crystal structures of E378D and R381M mutant enzymes have been determined to 1.7 and 2.1 A resolution, respectively. They allow an examination of the structural changes that disturb proton transport. Taken together, the results indicate that Arg-381, Glu-378, and Arg-402 form a proton pathway that is completely conserved throughout the fumarate reductase/succinate dehydrogenase family of enzymes.
Organizational Affiliation:
Edinburgh and St. Andrews Research School of Chemistry, University of Edinburgh, West Mains Road, Edinburgh EH9 3JJ, UK.