Structural basis for Rab11-dependent membrane recruitment of a family of Rab11-interacting protein 3 (FIP3)/Arfophilin-1.
Shiba, T., Koga, H., Shin, H.W., Kawasaki, M., Kato, R., Nakayama, K., Wakatsuki, S.(2006) Proc Natl Acad Sci U S A 103: 15416-15421
- PubMed: 17030804
- DOI: https://doi.org/10.1073/pnas.0605357103
- Primary Citation of Related Structures:
2D7C - PubMed Abstract:
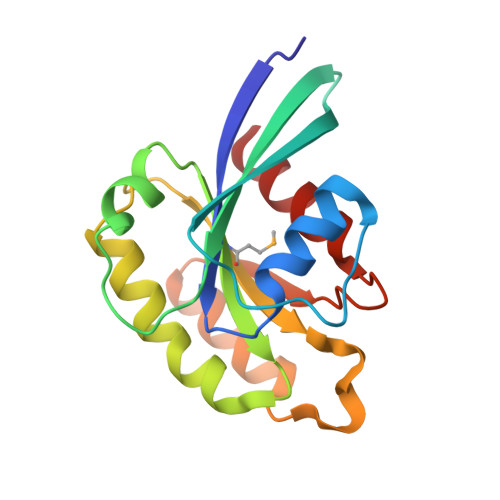
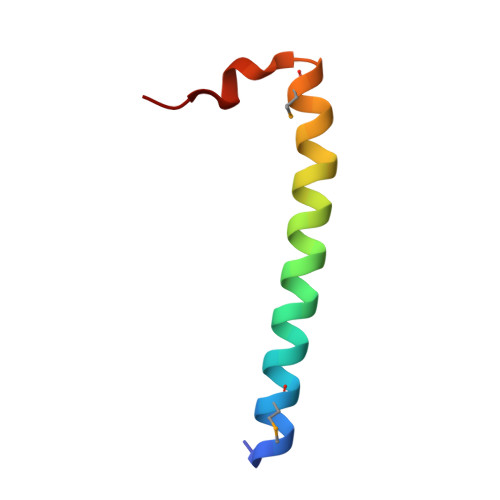
Family of Rab11-interacting protein (FIP)3/Arfophlin-1 and FIP4/Arfophilin-2 are dual effectors for Rab11 and ADP ribosylation factor (ARF)5/ARF6, which are involved in membrane delivery from recycling endosomes to the plasma membrane during cytokinesis. Here, we define the distinct C-terminal binding regions of FIP3 and FIP4 for Rab11 and ARF5/ARF6. Furthermore, we determined the crystal structure of Rab11 in complex with the Rab11-binding domain (RBD) of FIP3. The long amphiphilic alpha-helix of FIP3-RBD forms a parallel coiled-coil homodimer, with two symmetric interfaces with two Rab11 molecules. The hydrophobic side of the RBD helix is involved in homodimerization and mediates the interaction with the Rab11 switch 1 region, whereas the opposite hydrophilic side interacts with the Rab11 switch 2 and is the major factor contributing to the binding specificity. The bivalent interaction of FIP3 with Rab11 at the C terminus allows FIP3 to coordinately function with other binding partners, including ARFs.
- Structural Biology Research Center, Photon Factory, Institute of Materials Structure Science, High Energy Accelerator Research Organization, Tsukuba, Ibaraki 305-0801, Japan.
Organizational Affiliation: