Molecular Mechanism of Elongation Factor 1A Inhibition by a Legionella Pneumophila Glycosyltransferase.
Hurtado-Guerrero, R., Zusman, T., Pathak, S., Ibrahim, A.F.M., Shepherd, S., Prescott, A., Segal, G., Van Aalten, D.M.F.(2010) Biochem J 426: 281
- PubMed: 20030628
- DOI: https://doi.org/10.1042/BJ20091351
- Primary Citation of Related Structures:
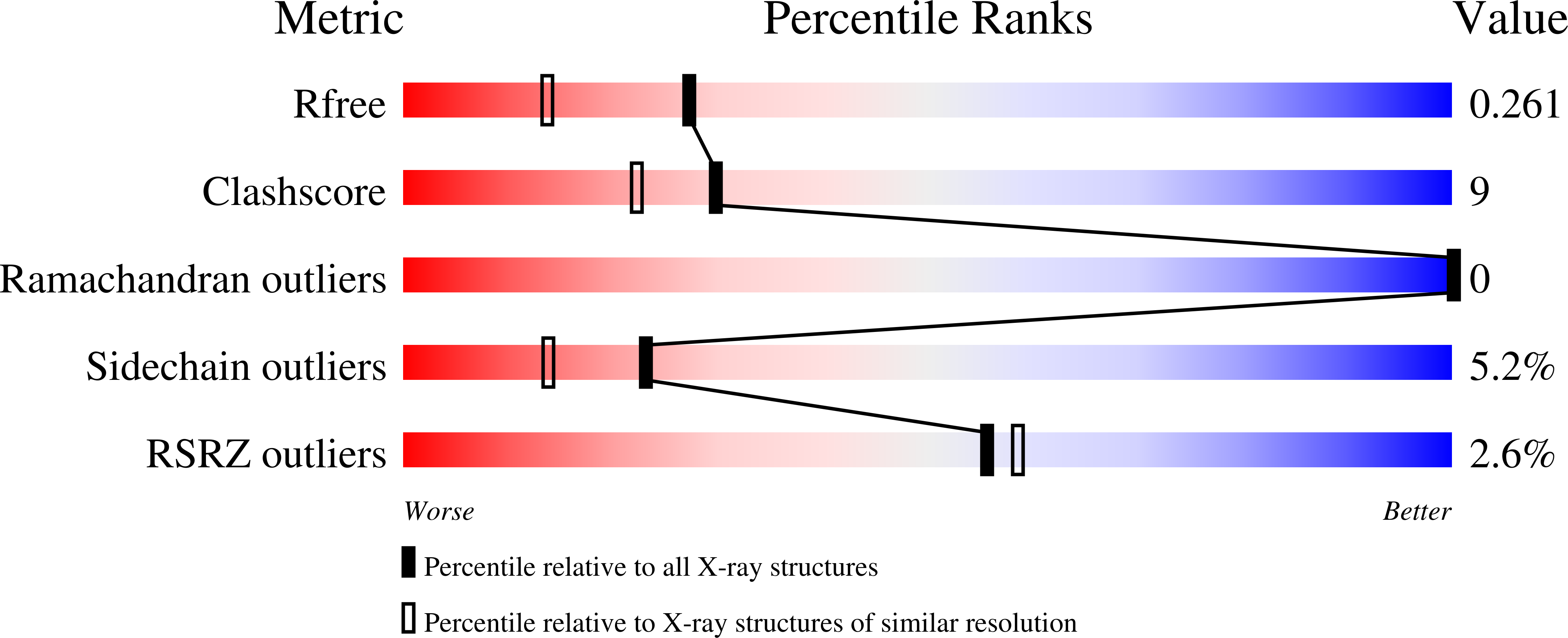
2WZF, 2WZG - PubMed Abstract:
Legionnaires' disease is caused by a lethal colonization of alveolar macrophages with the Gram-negative bacterium Legionella pneumophila. LpGT (L. pneumophila glucosyltransferase; also known as Lgt1) has recently been identified as a virulence factor, shutting down protein synthesis in the human cell by specific glucosylation of EF1A (elongation factor 1A), using an unknown mode of substrate recognition and a retaining mechanism for glycosyl transfer. We have determined the crystal structure of LpGT in complex with substrates, revealing a GT-A fold with two unusual protruding domains. Through structure-guided mutagenesis of LpGT, several residues essential for binding of the UDP-glucose-donor and EF1A-acceptor substrates were identified, which also affected L. pneumophila virulence as demonstrated by microinjection studies. Together, these results suggested that a positively charged EF1A loop binds to a negatively charged conserved groove on the LpGT structure, and that two asparagine residues are essential for catalysis. Furthermore, we showed that two further L. pneumophila glycosyltransferases possessed the conserved UDP-glucose-binding sites and EF1A-binding grooves, and are, like LpGT, translocated into the macrophage through the Icm/Dot (intracellular multiplication/defect in organelle trafficking) system.
Organizational Affiliation:
Division of Molecular Microbiology, College of Life Sciences, University of Dundee, Dundee DD1 5EH, Scotland, UK. R.HurtadoGuerrero@dundee.ac.uk