Structural basis for T cell alloreactivity among three HLA-B14 and HLA-B27 antigens
Kumar, P., Vahedi-Faridi, A., Saenger, W., Merino, E., Lopez de Castro, J.A., Uchanska-Ziegler, B., Ziegler, A.(2009) J Biol Chem 284: 29784-29797
- PubMed: 19617632
- DOI: https://doi.org/10.1074/jbc.M109.038497
- Primary Citation of Related Structures:
3BP4, 3BP7, 3BVN, 3BXN - PubMed Abstract:
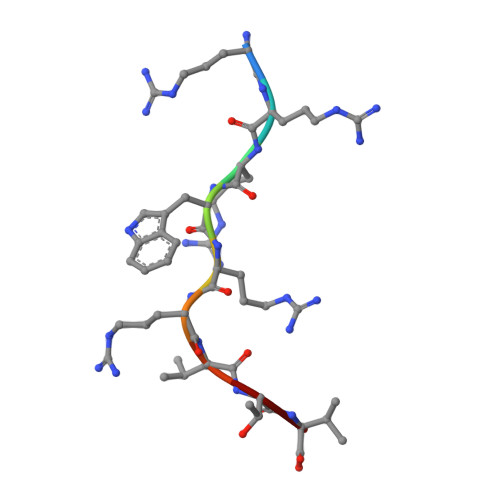
The existence of cytotoxic T cells (CTL) cross-reacting with the human major histocompatibility antigens HLA-B14 and HLA-B27 suggests that their alloreactivity could be due to presentation of shared peptides in similar binding modes by these molecules. We therefore determined the crystal structures of the subtypes HLA-B*1402, HLA-B*2705, and HLA-B*2709 in complex with a proven self-ligand, pCatA (peptide with the sequence IRAAPPPLF derived from cathepsin A (residues 2-10)), and of HLA-B*1402 in complex with a viral peptide, pLMP2 (RRRWRRLTV, derived from latent membrane protein 2 (residues 236-244) of Epstein-Barr virus). Despite the exchange of 18 residues within the binding grooves of HLA-B*1402 and HLA-B*2705 or HLA-B*2709, the pCatA peptide is presented in nearly identical conformations. However, pLMP2 is displayed by HLA-B*1402 in a conformation distinct from those previously found in the two HLA-B27 subtypes. In addition, the complexes of HLA-B*1402 with the two peptides reveal a nonstandard, tetragonal mode of the peptide N terminus anchoring in the binding groove because of the exchange of the common Tyr-171 by His-171 of the HLA-B*1402 heavy chain. This exchange appears also responsible for reduced stability of HLA-B14-peptide complexes in vivo and slow assembly in vitro. The studies with the pCatA peptide uncover that CTL cross-reactive between HLA-B14 and HLA-B27 might primarily recognize the common structural features of the bound peptide, thus neglecting amino acid replacements within the rim of the binding grooves. In contrast, structural alterations between the three complexes with the pLMP2 peptide indicate how heavy chain polymorphisms can influence peptide display and prevent CTL cross-reactivity between HLA-B14 and HLA-B27 antigens.
Organizational Affiliation:
Institut für Immungenetik, Charité-Universitätsmedizin Berlin, Campus Benjamin Franklin, Freie Universität Berlin, Thielallee 73, 14195 Berlin, Germany.