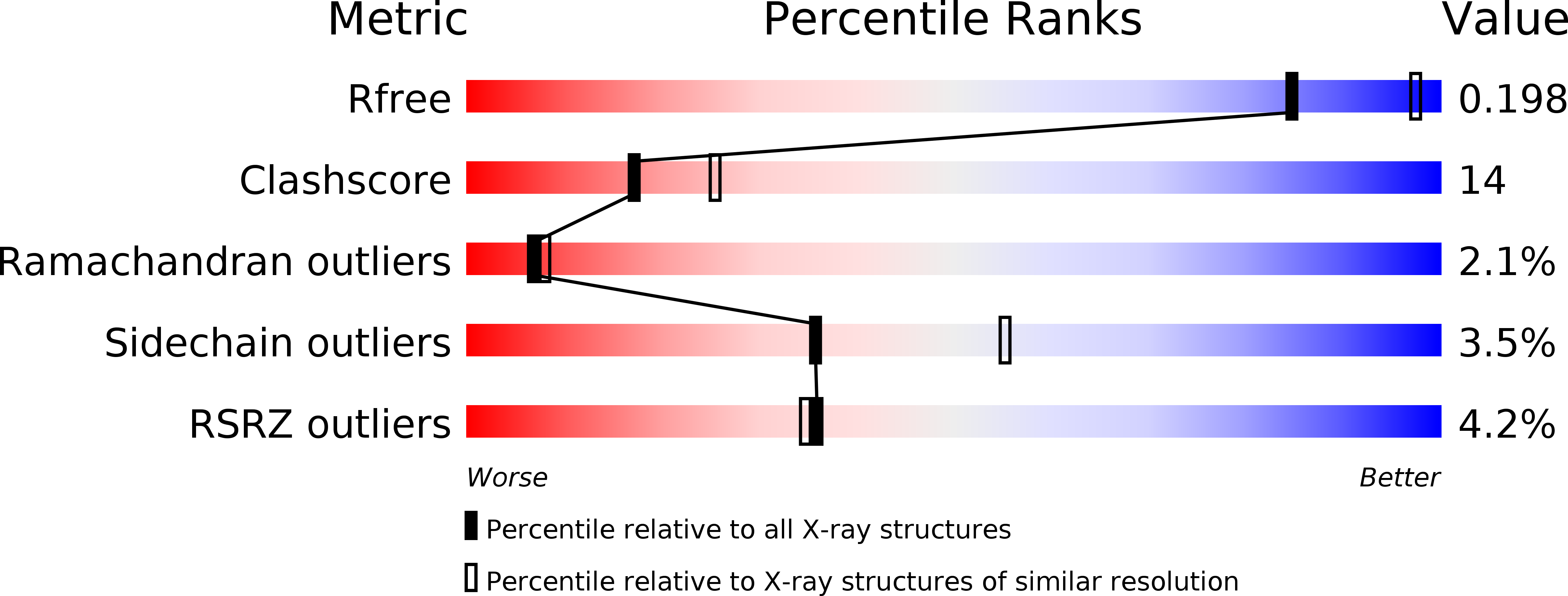
Crystal structures of a therapeutic single chain antibody in complex with two drugs of abuse-Methamphetamine and 3,4-methylenedioxymethamphetamine.
Celikel, R., Peterson, E.C., Owens, S.M., Varughese, K.I.(2009) Protein Sci 18: 2336-2345
- PubMed: 19760665
- DOI: https://doi.org/10.1002/pro.244
- Primary Citation of Related Structures:
3GKZ, 3GM0 - PubMed Abstract:
Methamphetamine (METH) is a major drug threat in the United States and worldwide. Monoclonal antibody (mAb) therapy for treating METH abuse is showing exciting promise and the understanding of how mAb structure relates to function will be essential for future development of these important therapies. We have determined crystal structures of a high affinity anti-(+)-METH therapeutic single chain antibody fragment (scFv6H4, K(D)= 10 nM) derived from one of our candidate mAb in complex with METH and the (+) stereoisomer of another abused drug, 3,4-methylenedioxymethamphetamine (MDMA), known by the street name "ecstasy." The crystal structures revealed that scFv6H4 binds to METH and MDMA in a deep pocket that almost completely encases the drugs mostly through aromatic interactions. In addition, the cationic nitrogen of METH and MDMA forms a salt bridge with the carboxylate group of a glutamic acid residue and a hydrogen bond with a histidine side chain. Interestingly, there are two water molecules in the binding pocket and one of them is positioned for a C--H...O interaction with the aromatic ring of METH. These first crystal structures of a high affinity therapeutic antibody fragment against METH and MDMA (resolution = 1.9 A, and 2.4 A, respectively) provide a structural basis for designing the next generation of higher affinity antibodies and also for carrying out rational humanization.
Organizational Affiliation:
Department of Physiology and Biophysics, College of Medicine, University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA.