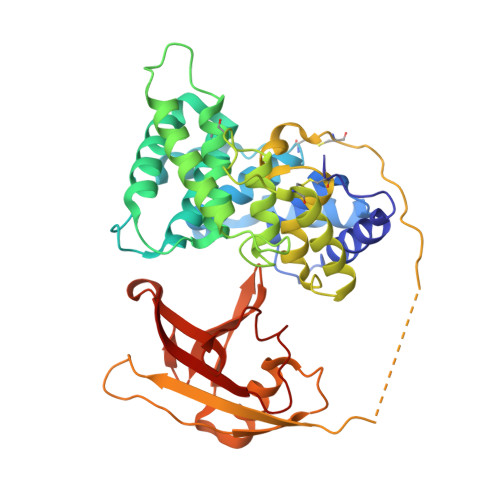
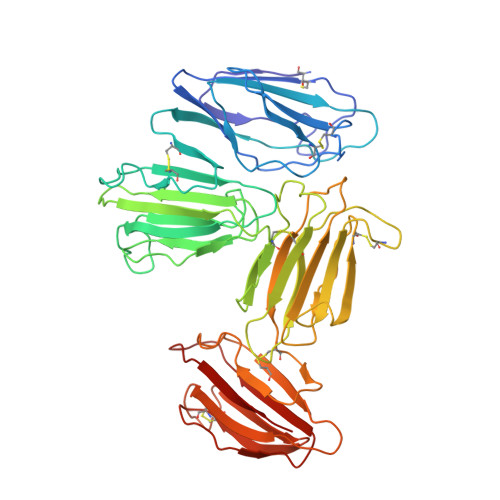
Structural basis for receptor recognition of vitamin-B(12)-intrinsic factor complexes.
Andersen, C.B., Madsen, M., Storm, T., Moestrup, S.K., Andersen, G.R.(2010) Nature 464: 445-448
- PubMed: 20237569
- DOI: https://doi.org/10.1038/nature08874
- Primary Citation of Related Structures:
3KQ4 - PubMed Abstract:
Cobalamin (Cbl, vitamin B(12)) is a bacterial organic compound and an essential coenzyme in mammals, which take it up from the diet. This occurs by the combined action of the gastric intrinsic factor (IF) and the ileal endocytic cubam receptor formed by the 460-kilodalton (kDa) protein cubilin and the 45-kDa transmembrane protein amnionless. Loss of function of any of these proteins ultimately leads to Cbl deficiency in man. Here we present the crystal structure of the complex between IF-Cbl and the cubilin IF-Cbl-binding-region (CUB(5-8)) determined at 3.3 A resolution. The structure provides insight into how several CUB (for 'complement C1r/C1s, Uegf, Bmp1') domains collectively function as modular ligand-binding regions, and how two distant CUB domains embrace the Cbl molecule by binding the two IF domains in a Ca(2+)-dependent manner. This dual-point model provides a probable explanation of how Cbl indirectly induces ligand-receptor coupling. Finally, the comparison of Ca(2+)-binding CUB domains and the low-density lipoprotein (LDL) receptor-type A modules suggests that the electrostatic pairing of a basic ligand arginine/lysine residue with Ca(2+)-coordinating acidic aspartates/glutamates is a common theme of Ca(2+)-dependent ligand-receptor interactions.
Organizational Affiliation:
Department of Medical Biochemistry, Aarhus University, 8000 Aarhus C, Denmark.