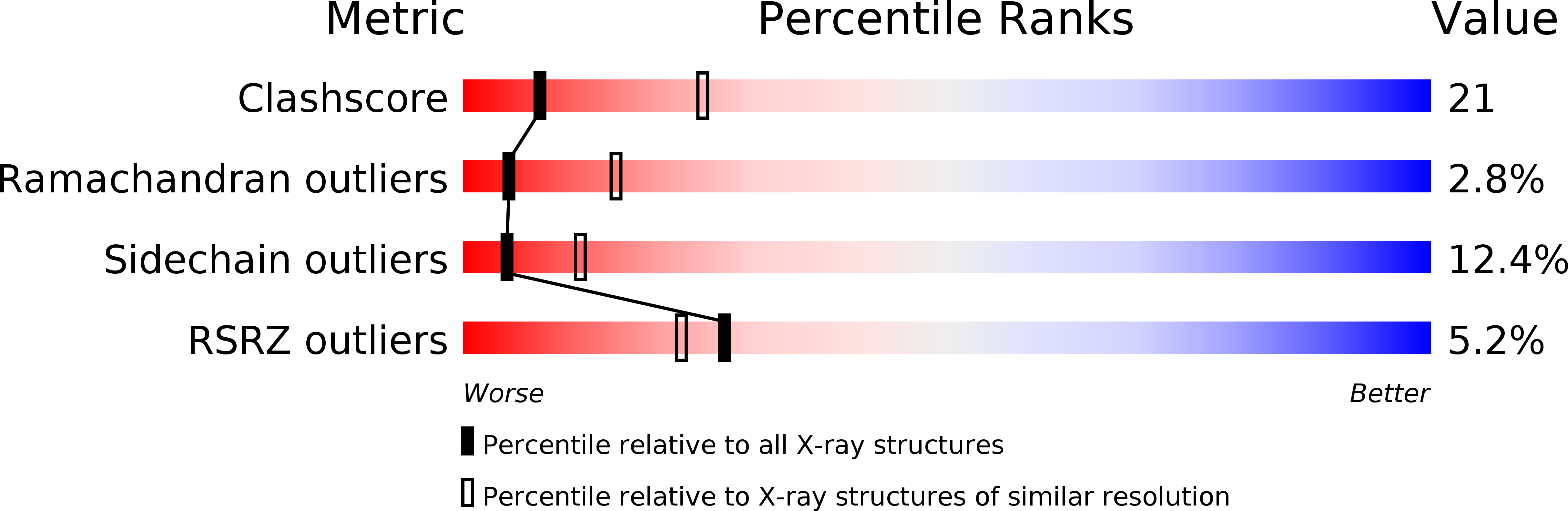
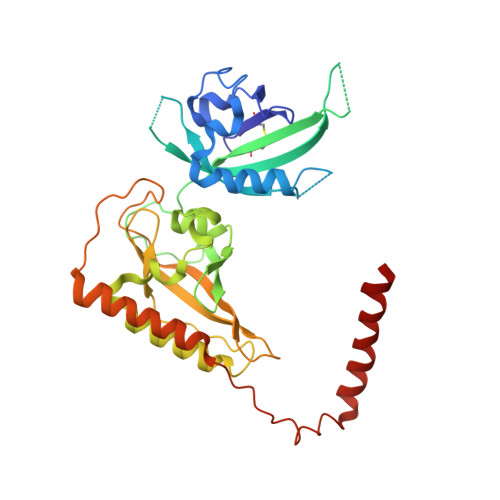
Structure of an enclosed dimer formed by the Drosophila period protein.
King, H.A., Hoelz, A., Crane, B.R., Young, M.W.(2011) J Mol Biol 413: 561-572
- PubMed: 21907720
- DOI: https://doi.org/10.1016/j.jmb.2011.08.048
- Primary Citation of Related Structures:
3RTY - PubMed Abstract:
Period (PER) is the major transcription inhibitor in metazoan circadian clocks and lies at the center of several feedback loops that regulate gene expression. Dimerization of Drosophila PER influences nuclear translocation, repressor activity, and behavioral rhythms. The structure of a central, 346-residue PER fragment reveals two associated PAS (Per-Arnt-Sim) domains followed by a protruding α-helical extension (αF). A closed, pseudo-symmetric dimer forms from a cross handshake interaction of the N-terminal PAS domain with αF of the opposing subunit. Strikingly, a shift of αF against the PAS β-sheet generates two alternative subunit interfaces in the dimer. Taken together with a previously reported PER structure in which αF extends, these data indicate that αF unlatches to switch association of PER with itself to its partner Timeless. The variable positions of the αF helix provide snapshots of a helix dissociation mechanism that has relevance to other PAS protein systems. Conservation of PER interaction residues among a family of PAS-AB-containing transcription factors suggests that contacts mediating closed PAS-AB dimers serve a general function.
Organizational Affiliation:
Laboratory of Genetics, The Rockefeller University, New York, NY 10065, USA.