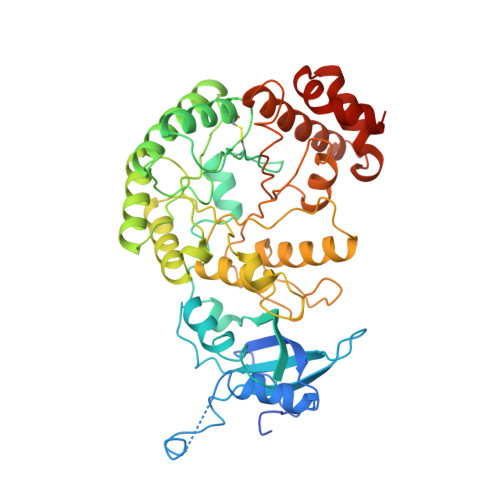
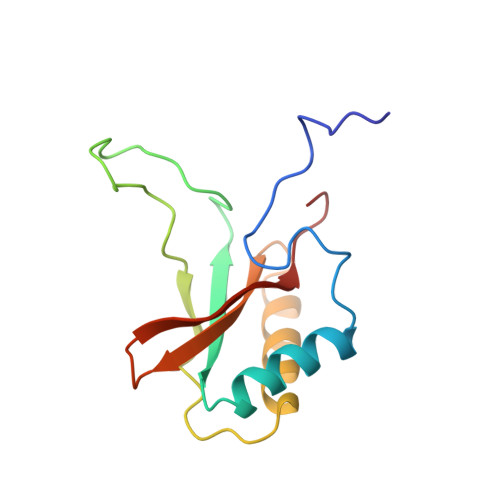
Crystal structure of the unactivated form of ribulose-1,5-bisphosphate carboxylase/oxygenase from tobacco refined at 2.0-A resolution.
Curmi, P.M., Cascio, D., Sweet, R.M., Eisenberg, D., Schreuder, H.(1992) J Biol Chem 267: 16980-16989
- PubMed: 1512238
- Primary Citation of Related Structures:
3RUB - PubMed Abstract:
The structure of the unactivated form of ribulose-1,5-bisphosphate carboxylase/oxygenase was refined at a resolution of 2.0 A to an R-factor of 17.1%. The previous model (Chapman et al., 1988) was extensively rebuilt, and the small subunit was retraced. The refined model consists of residues 22-63 and 69-467 of the large subunit and the complete small subunit. A striking feature of the model is that several loops have very high B-factors, probably representing mobile regions of the molecule. An examination of the intersubunit contacts shows that the L8S8 hexadecamer is composed of four L2 dimers. The dominant contacts between these L2 dimers are formed by the small subunits. This suggests that the small subunits may be essential for maintaining the integrity of the L8S8 structure. The active site shows differences between the unactivated form and the quaternary complex. In particular, Lys334 has moved out of the active site by about 10A. This residue lies on loop 6 of the alpha beta barrel, which is a particularly mobile loop. The site of ribulose-1,5-bisphosphate carboxylase/oxygenase activation is well ordered in the absence of the carbamylation of Lys201 and Mg2+ binding. The residues are held poised by a network of hydrogen bonds. In the unactivated state, the active site is accessible to substrate binding.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, Los Angeles 90024.