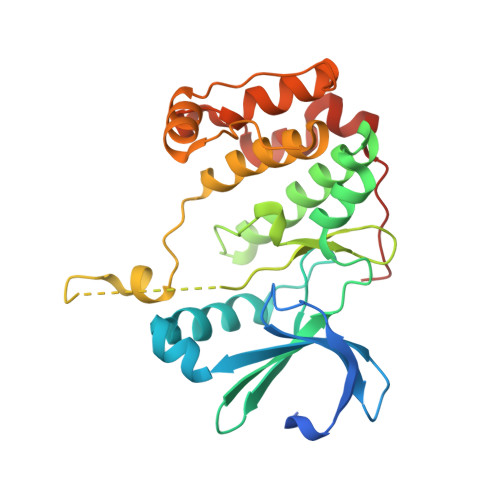
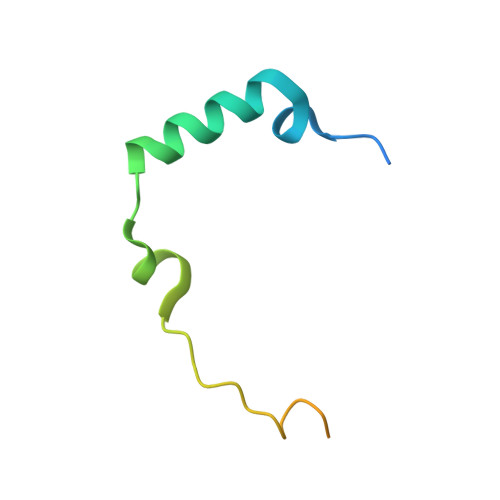
Crystal Structure of Human Aurora B in Complex with Incenp and Vx-680.
Elkins, J.M., Santaguida, S., Musacchio, A., Knapp, S.(2012) J Med Chem 55: 7841
- PubMed: 22920039
- DOI: https://doi.org/10.1021/jm3008954
- Primary Citation of Related Structures:
4AF3 - PubMed Abstract:
We present the structure of the human Aurora B kinase domain in complex with the C-terminal Aurora-binding region of human INCENP and the Aurora kinase inhibitor VX-680. The structure unexpectedly reveals a dimeric arrangement of the Aurora B:INCENP complex, which was confirmed to exist in solution by analytical ultracentrifugation. The dimerization involves a domain swap of the activation loop, resulting in a different conformation of the DFG motif as compared to that seen in other kinase complexes with VX-680. The binding of INCENP differs significantly from that seen in the Xenopus laevis Aurora B:INCENP complex currently used as a model for structure-based design for this important oncology target.
Organizational Affiliation:
Nuffield Department of Clinical Medicine, Structural Genomics Consortium, University of Oxford , Old Road Campus Research Building, Roosevelt Drive, Oxford, OX3 7DQ, United Kingdom. jon.elkins@sgc.ox.ac.uk