Contamination from an affinity column: an encounter with a new villain in the world of membrane-protein crystallization.
Panwar, P., Deniaud, A., Pebay-Peyroula, E.(2012) Acta Crystallogr D Biol Crystallogr 68: 1272-1277
- PubMed: 22993081
- DOI: https://doi.org/10.1107/S090744491202639X
- Primary Citation of Related Structures:
4DNE - PubMed Abstract:
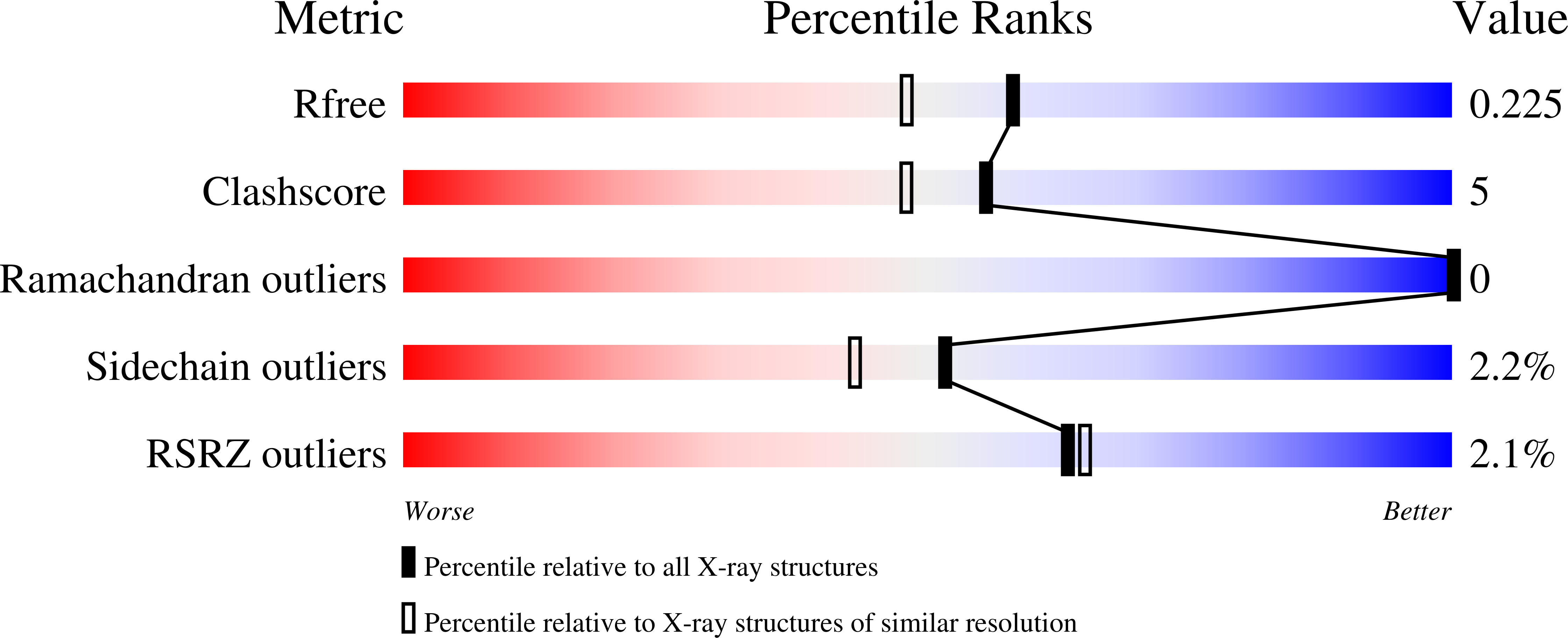
Attempts to crystallize AtNTT1, a chloroplast ATP/ADP transporter from Arabidopsis thaliana, revealed an unexpected contaminant, Strep-Tactin, a variant of streptavidin that was used during purification of the protein. Although it was present in very small amounts, crystals of Strep-Tactin were reproducibly grown from the AtNTT1 solution. AtNTT1 was overexpressed in Escherichia coli and purified from detergent-solubilized membrane fractions using Strep-Tactin affinity chromatography based on an engineered streptavidin. The contamination of protein solutions purified on Strep-Tactin columns has never been described previously and seems to be specific to membrane proteins solubilized in detergents. Trace amounts of Strep-Tactin were observed to be eluted from a Strep-Tactin column using several routinely used detergents, illustrating their possible role in the contamination. This finding raises an alarm and suggests caution in membrane-protein purification using Strep-Tactin affinity columns, where detergents are essential components. The small crystals of contaminant protein led to the structure at 1.9 Å resolution of Strep-Tactin in complex with desthiobiotin.
Organizational Affiliation:
CEA, Institut de Biologie Structurale Jean-Pierre Ebel, 41 Rue Jules Horowitz, 38027 Grenoble, France.