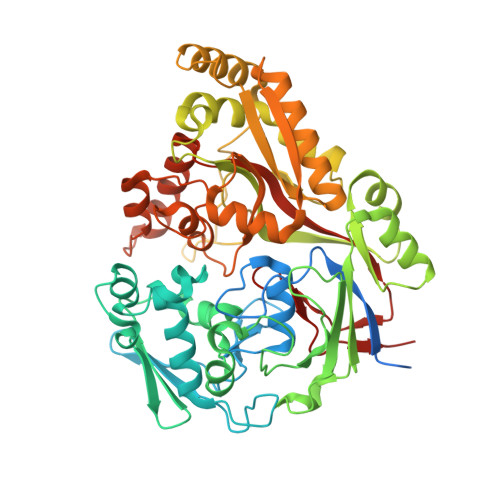
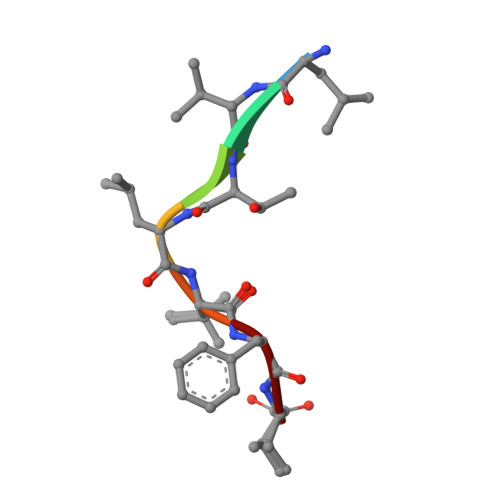
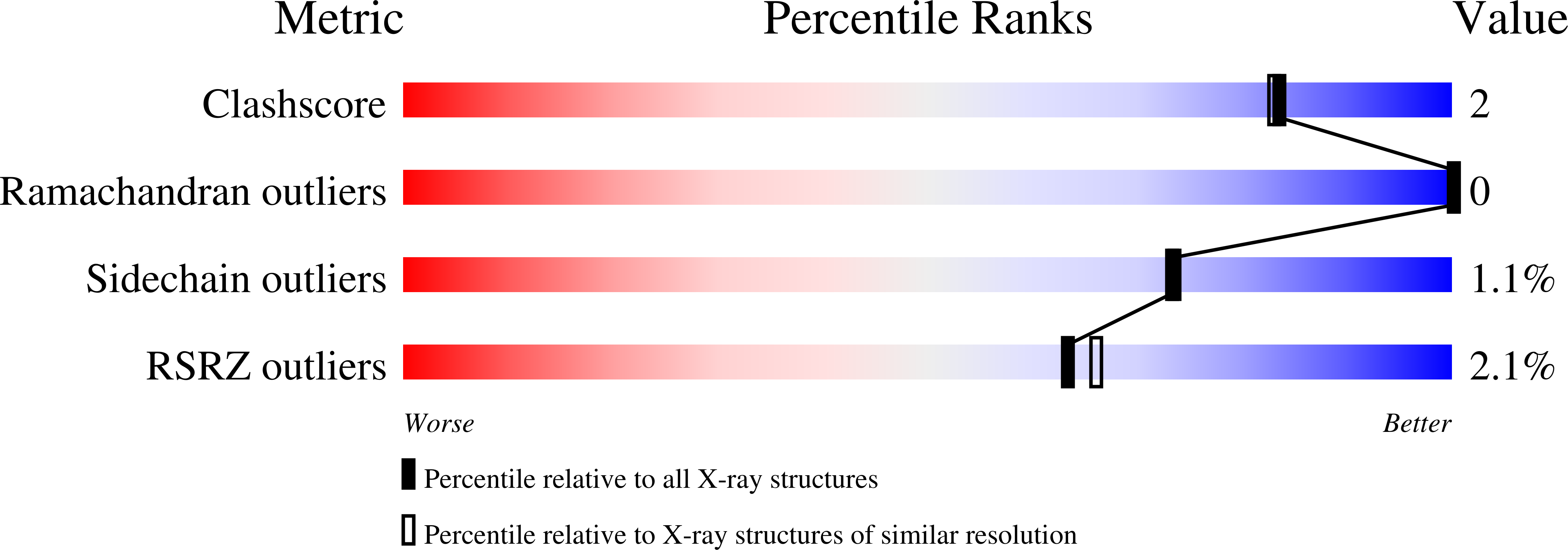Structure and Mode of Peptide Binding of Pheromone Receptor PrgZ.
Berntsson, R.P., Schuurman-Wolters, G.K., Dunny, G., Slotboom, D.J., Poolman, B.(2012) J Biol Chem 287: 37165-37170
- PubMed: 22948145
- DOI: https://doi.org/10.1074/jbc.M112.386334
- Primary Citation of Related Structures:
4FAJ - PubMed Abstract:
We present the crystal structure of the pheromone receptor protein PrgZ from Enterococcus faecalis in complex with the heptapeptide cCF10 (LVTLVFV), which is used in signaling between conjugative recipient and donor cells. Comparison of PrgZ with homologous oligopeptide-binding proteins (AppA and OppA) explains the high specificity of PrgZ for hydrophobic heptapeptides versus the promiscuity of peptide binding in the homologous proteins.
Organizational Affiliation:
Department of Biochemistry, Groningen Biomolecular Sciences and Biotechnology Institute, Netherlands Proteomics Centre, University of Groningen, 9747 AG Groningen, The Netherlands.