Structural Basis for the Recognition of Peptide RJPXD33 by Acyltransferases in Lipid A Biosynthesis.
Jenkins, R.J., Heslip, K.A., Meagher, J.L., Stuckey, J.A., Dotson, G.D.(2014) J Biological Chem 289: 15527-15535
- PubMed: 24742680
- DOI: https://doi.org/10.1074/jbc.M114.564278
- Primary Citation of Related Structures:
4J09 - PubMed Abstract:
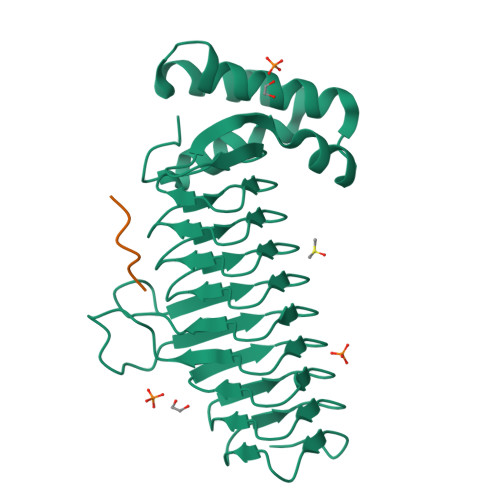
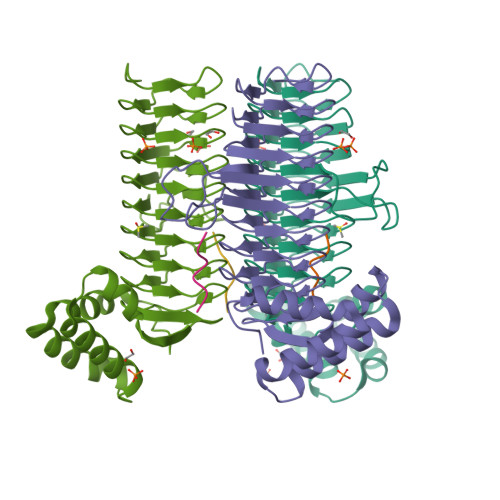
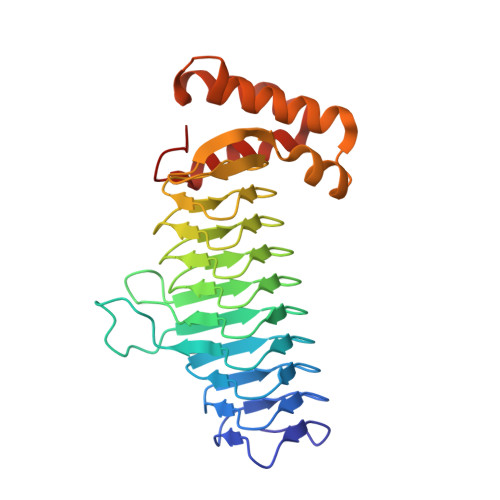
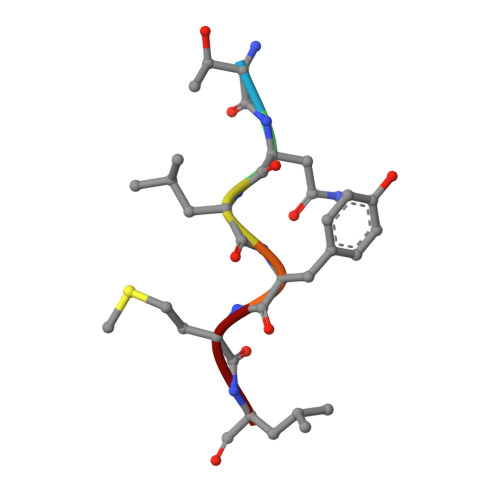
UDP-N-acetylglucosamine acyltransferase (LpxA) and UDP-3-O-(acyl)-glucosamine acyltransferase (LpxD) constitute the essential, early acyltransferases of lipid A biosynthesis. Recently, an antimicrobial peptide inhibitor, RJPXD33, was identified with dual affinity for LpxA and LpxD. To gain a fundamental understanding of the molecular basis of inhibitor binding, we determined the crystal structure of LpxA from Escherichia coli in complex with RJPXD33 at 1.9 Å resolutions. Our results suggest that the peptide binds in a unique modality that mimics (R)-β-hydroxyacyl pantetheine binding to LpxA and displays how the peptide binds exclusive of the native substrate, acyl-acyl carrier protein. Acyltransferase binding studies with photo-labile RJPXD33 probes and truncations of RJPXD33 validated the structure and provided fundamental insights for future design of small molecule inhibitors. Overlay of the LpxA-RJPXD33 structure with E. coli LpxD identified a complementary peptide binding pocket within LpxD and serves as a model for further biochemical characterization of RJPXD33 binding to LpxD.
Organizational Affiliation:
From the Department of Medicinal Chemistry, College of Pharmacy, and.