Ribosome Biogenesis Factor Tsr3 is the Aminocarboxypropyl Transferase Responsible for 18S Rrna Hypermodification in Yeast and Humans
Entian, K.-D., Immer, C., Koetter, P., Lafontaine, D., Britter, M., Pogoryelov, D., Sharma, S., Wohnert, J., Wurm, J.P.(2016) Nucleic Acids Res 44: 4304
- PubMed: 27084949
- DOI: https://doi.org/10.1093/nar/gkw244
- Primary Citation of Related Structures:
5AP8, 5APG - PubMed Abstract:
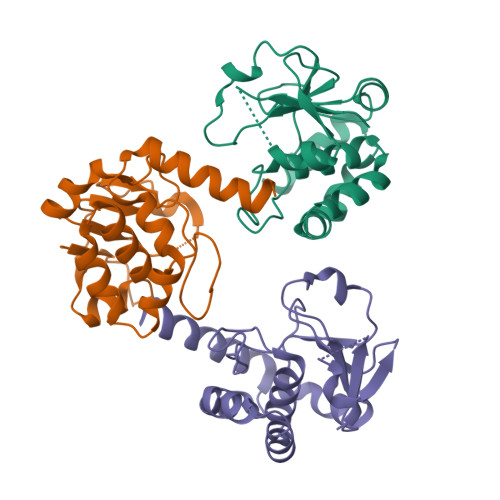
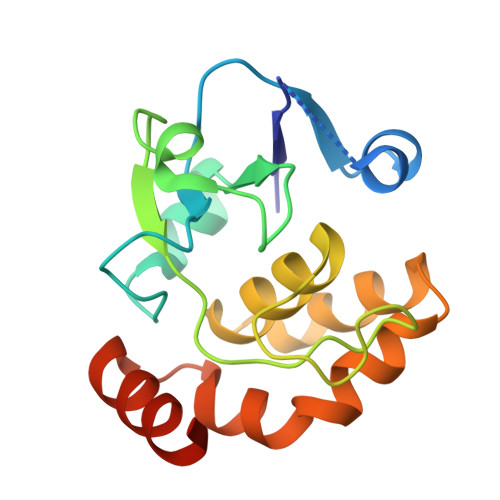
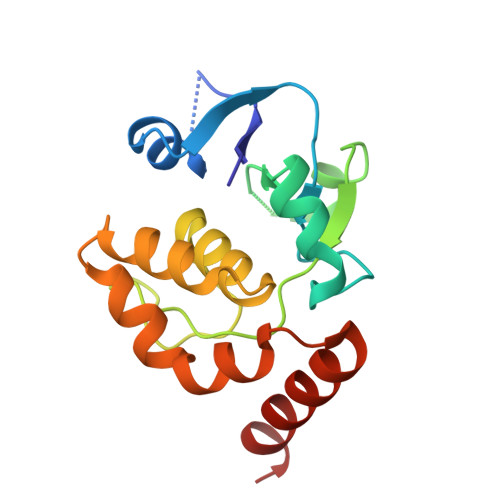
The chemically most complex modification in eukaryotic rRNA is the conserved hypermodified nucleotide N1-methyl-N3-aminocarboxypropyl-pseudouridine (m(1)acp(3)Ψ) located next to the P-site tRNA on the small subunit 18S rRNA. While S-adenosylmethionine was identified as the source of the aminocarboxypropyl (acp) group more than 40 years ago the enzyme catalyzing the acp transfer remained elusive. Here we identify the cytoplasmic ribosome biogenesis protein Tsr3 as the responsible enzyme in yeast and human cells. In functionally impaired Tsr3-mutants, a reduced level of acp modification directly correlates with increased 20S pre-rRNA accumulation. The crystal structure of archaeal Tsr3 homologs revealed the same fold as in SPOUT-class RNA-methyltransferases but a distinct SAM binding mode. This unique SAM binding mode explains why Tsr3 transfers the acp and not the methyl group of SAM to its substrate. Structurally, Tsr3 therefore represents a novel class of acp transferase enzymes.
Organizational Affiliation:
Institute for Molecular Biosciences, Goethe University, Frankfurt/M, Germany.