Design, synthesis and biological evaluation of new peptidomimetic compounds targeting NRP-1 receptor TO BE PUBLISHED
Richard, M., Pelligrini Moise, N., Jelsch, C., Maigret, B., Didierjean, C., Manival, X., Charron, C.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Neuropilin-1 | 161 | Homo sapiens | Mutation(s): 0 Gene Names: NRP1, NRP, VEGF165R | 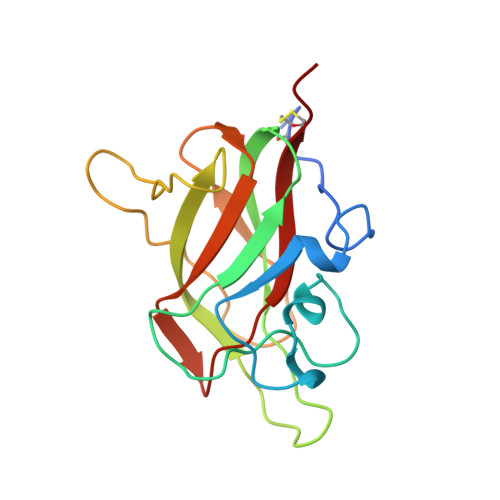 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O14786 (Homo sapiens) Explore O14786 Go to UniProtKB: O14786 | |||||
PHAROS: O14786 GTEx: ENSG00000099250 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O14786 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| BCN Query on BCN | B [auth A] | BICINE C6 H13 N O4 FSVCELGFZIQNCK-UHFFFAOYSA-N |  | ||
| NA Query on NA | C [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 62.28 | α = 90 |
| b = 62.28 | β = 90 |
| c = 85.96 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| Coot | model building |