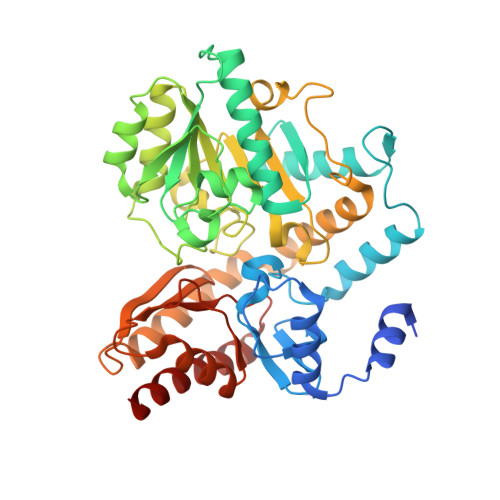
Structure of the ornithine aminotransferase from Toxoplasma gondii in complex with inactivator.
Filippova, E.V., Minasov, G., Flores, K., Le, H.V., Silverman, R.B., McLeod, R.L., Anderson, W.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.