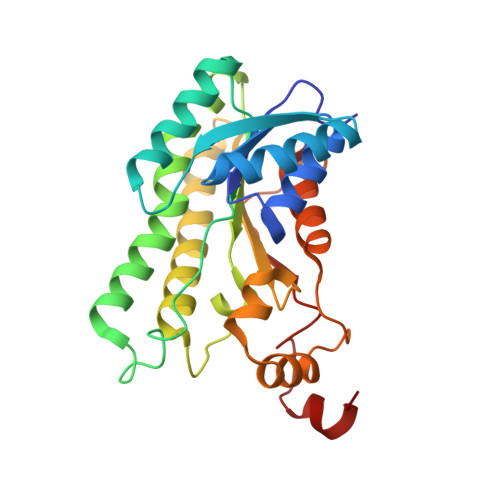
The three-dimensional structure of Clostridium absonum 7 alpha-hydroxysteroid dehydrogenase: new insights into the conserved arginines for NADP(H) recognition
Lou, D., Wang, B., Tan, J., Zhu, L., Cen, X., Ji, Q., Wang, Y.(2016) Sci Rep 6: 22885-22885
- PubMed: 26961171
- DOI: https://doi.org/10.1038/srep22885
- Primary Citation of Related Structures:
5EPO - PubMed Abstract:
7α-hydroxysteroid dehydrogenase (7α-HSDH) can catalyse the oxidation of C7 α-OH of the steroid nucleus in the bile acid metabolism. In the paper we determined the crystal structure of 7α-HSDH from Clostridium absonum (CA 7α-HSDH) complexed with taurochenodeoxycholic acid (TCDCA) and NADP(+) by X-ray diffraction, which, as a tetramer, possesses the typical α/β folding pattern. The four subunits of an asymmetric unit lie in the fact that there are the stable hydrophobic interactions between Q-axis-related subunits. Significantly, we captured an active state of the NADP(+), confirming that nicotinamide moiety of NADP(+) act as electron carrier in the dehydrogenation. On the basis of crystal structure analysis, site-directed mutagenesis and MD simulation, furthermore, we find that the guanidinium of Arg38 can form the stable cation-π interaction with the adenine ring of NADP(+), and the cation-π interaction and hydrogen bonds between Arg38 and NADP(+) have a significant anchor effect on the cofactor binding to CA 7α-HSDH.
Organizational Affiliation:
Key Laboratory of Biorheological Science and Technology (Chongqing University), Ministry of Education, College of Bioengineering, Chongqing University, Chongqing 400030, China.