Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Lin, H., Zeng, J., Xie, R., Schulz, M.J., Tedesco, R., Qu, J., Erhard, K.F., Mack, J.F., Raha, K., Rendina, A.R., Szewczuk, L.M., Kratz, P.M., Jurewicz, A.J., Cecconie, T., Martens, S., McDevitt, P.J., Martin, J.D., Chen, S.B., Jiang, Y., Nickels, L., Schwartz, B.J., Smallwood, A., Zhao, B., Campobasso, N., Qian, Y., Briand, J., Rominger, C.M., Oleykowski, C., Hardwicke, M.A., Luengo, J.I.(2016) ACS Med Chem Lett 7: 217-222
- PubMed: 26985301
- DOI: https://doi.org/10.1021/acsmedchemlett.5b00214
- Primary Citation of Related Structures:
5HEX, 5HFU, 5HG1 - PubMed Abstract:
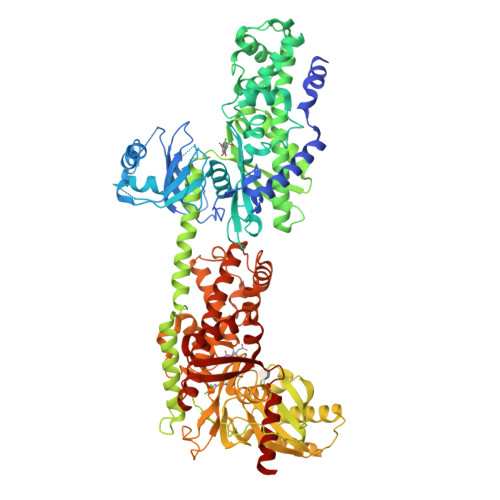
A novel series of potent and selective hexokinase 2 (HK2) inhibitors, 2,6-disubstituted glucosamines, has been identified based on HTS hits, exemplified by compound 1. Inhibitor-bound crystal structures revealed that the HK2 enzyme could adopt an "induced-fit" conformation. The SAR study led to the identification of potent HK2 inhibitors, such as compound 34 with greater than 100-fold selectivity over HK1. Compound 25 inhibits in situ glycolysis in a UM-UC-3 bladder tumor cell line via (13)CNMR measurement of [3-(13)C]lactate produced from [1,6-(13)C2]glucose added to the cell culture.
Organizational Affiliation:
Cancer Metabolism Chemistry; Cancer Metabolism Biology; and Platform Technology & Sciences, GlaxoSmithKline , 1250 South Collegeville Road, Collegeville, Pennsylvania 19426-0989, United States.