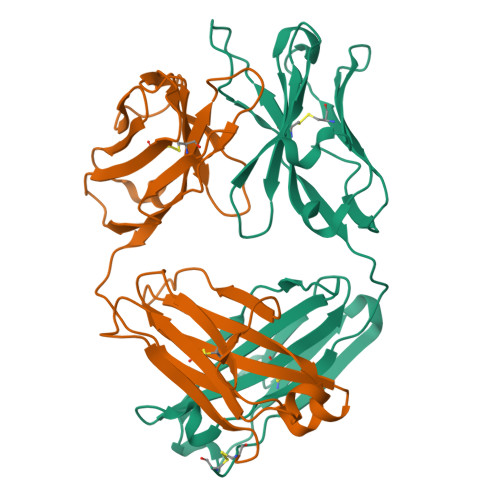
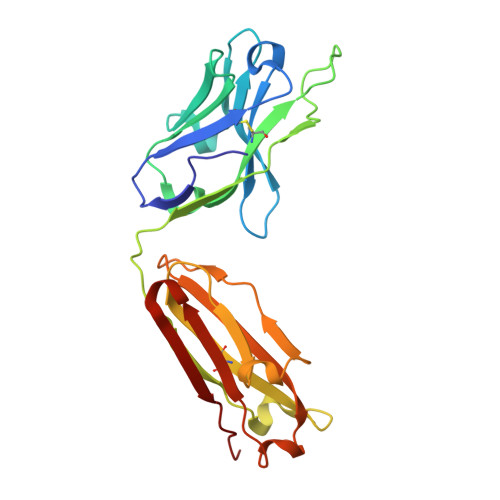
Structural basis for nonneutralizing antibody competition at antigenic site II of the respiratory syncytial virus fusion protein.
Mousa, J.J., Sauer, M.F., Sevy, A.M., Finn, J.A., Bates, J.T., Alvarado, G., King, H.G., Loerinc, L.B., Fong, R.H., Doranz, B.J., Correia, B.E., Kalyuzhniy, O., Wen, X., Jardetzky, T.S., Schief, W.R., Ohi, M.D., Meiler, J., Crowe, J.E.(2016) Proc Natl Acad Sci U S A 113: E6849-E6858
- PubMed: 27791117
- DOI: https://doi.org/10.1073/pnas.1609449113
- Primary Citation of Related Structures:
5ITB, 5J3D - PubMed Abstract:
Palivizumab was the first antiviral monoclonal antibody (mAb) approved for therapeutic use in humans, and remains a prophylactic treatment for infants at risk for severe disease because of respiratory syncytial virus (RSV). Palivizumab is an engineered humanized version of a murine mAb targeting antigenic site II of the RSV fusion (F) protein, a key target in vaccine development. There are limited reported naturally occurring human mAbs to site II; therefore, the structural basis for human antibody recognition of this major antigenic site is poorly understood. Here, we describe a nonneutralizing class of site II-specific mAbs that competed for binding with palivizumab to postfusion RSV F protein. We also describe two classes of site II-specific neutralizing mAbs, one of which escaped competition with nonneutralizing mAbs. An X-ray crystal structure of the neutralizing mAb 14N4 in complex with F protein showed that the binding angle at which human neutralizing mAbs interact with antigenic site II determines whether or not nonneutralizing antibodies compete with their binding. Fine-mapping studies determined that nonneutralizing mAbs that interfere with binding of neutralizing mAbs recognize site II with a pose that facilitates binding to an epitope containing F surface residues on a neighboring protomer. Neutralizing antibodies, like motavizumab and a new mAb designated 3J20 that escape interference by the inhibiting mAbs, avoid such contact by binding at an angle that is shifted away from the nonneutralizing site. Furthermore, binding to rationally and computationally designed site II helix-loop-helix epitope-scaffold vaccines distinguished neutralizing from nonneutralizing site II antibodies.
Organizational Affiliation:
Vanderbilt Vaccine Center, Vanderbilt University Medical Center, Nashville, TN 37232.