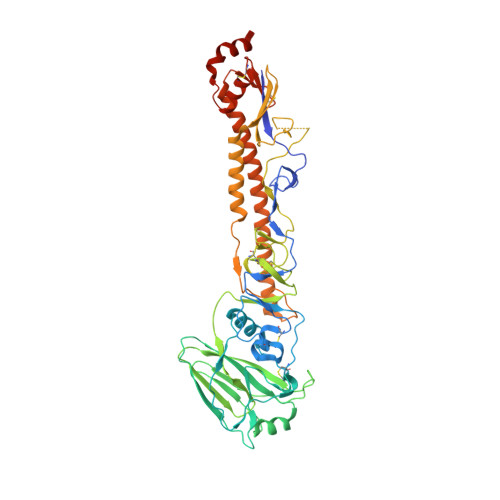
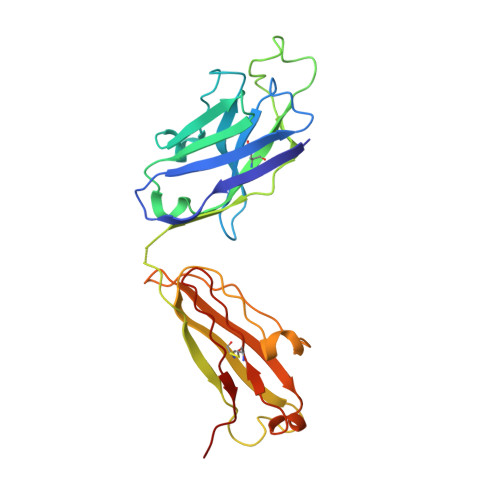
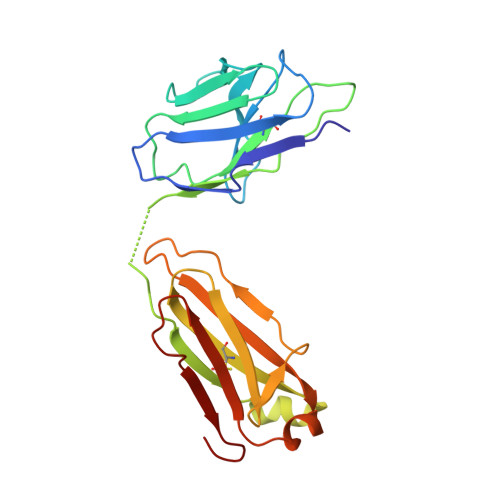
Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses.
Joyce, M.G., Wheatley, A.K., Thomas, P.V., Chuang, G.Y., Soto, C., Bailer, R.T., Druz, A., Georgiev, I.S., Gillespie, R.A., Kanekiyo, M., Kong, W.P., Leung, K., Narpala, S.N., Prabhakaran, M.S., Yang, E.S., Zhang, B., Zhang, Y., Asokan, M., Boyington, J.C., Bylund, T., Darko, S., Lees, C.R., Ransier, A., Shen, C.H., Wang, L., Whittle, J.R., Wu, X., Yassine, H.M., Santos, C., Matsuoka, Y., Tsybovsky, Y., Baxa, U., Mullikin, J.C., Subbarao, K., Douek, D.C., Graham, B.S., Koup, R.A., Ledgerwood, J.E., Roederer, M., Shapiro, L., Kwong, P.D., Mascola, J.R., McDermott, A.B.(2016) Cell 166: 609-623
- PubMed: 27453470
- DOI: https://doi.org/10.1016/j.cell.2016.06.043
- Primary Citation of Related Structures:
5K9J, 5K9K, 5K9O, 5K9Q, 5KAN, 5KAQ - PubMed Abstract:
Antibodies capable of neutralizing divergent influenza A viruses could form the basis of a universal vaccine. Here, from subjects enrolled in an H5N1 DNA/MIV-prime-boost influenza vaccine trial, we sorted hemagglutinin cross-reactive memory B cells and identified three antibody classes, each capable of neutralizing diverse subtypes of group 1 and group 2 influenza A viruses. Co-crystal structures with hemagglutinin revealed that each class utilized characteristic germline genes and convergent sequence motifs to recognize overlapping epitopes in the hemagglutinin stem. All six analyzed subjects had sequences from at least one multidonor class, and-in half the subjects-multidonor-class sequences were recovered from >40% of cross-reactive B cells. By contrast, these multidonor-class sequences were rare in published antibody datasets. Vaccination with a divergent hemagglutinin can thus increase the frequency of B cells encoding broad influenza A-neutralizing antibodies. We propose the sequence signature-quantified prevalence of these B cells as a metric to guide universal influenza A immunization strategies.
Organizational Affiliation:
Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892, USA.