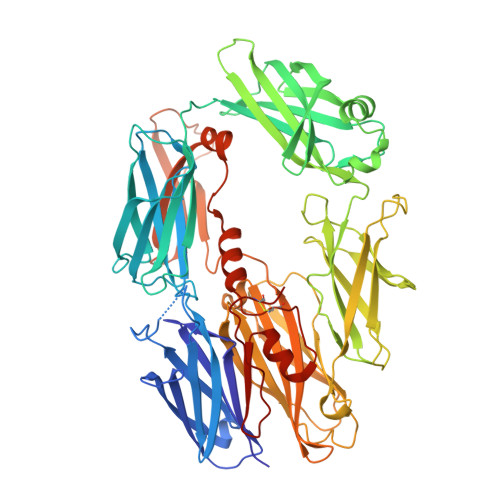
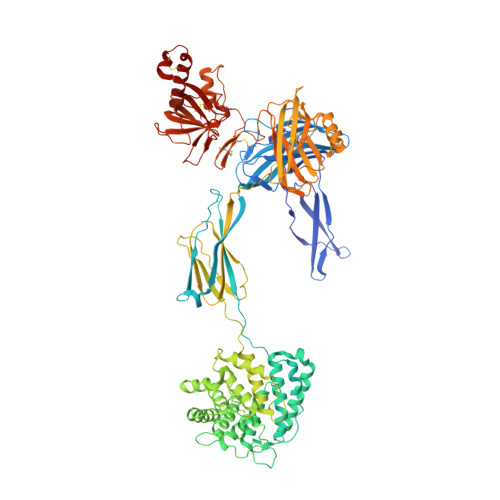
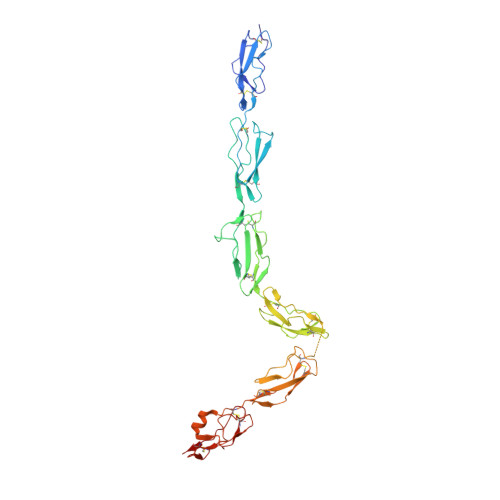
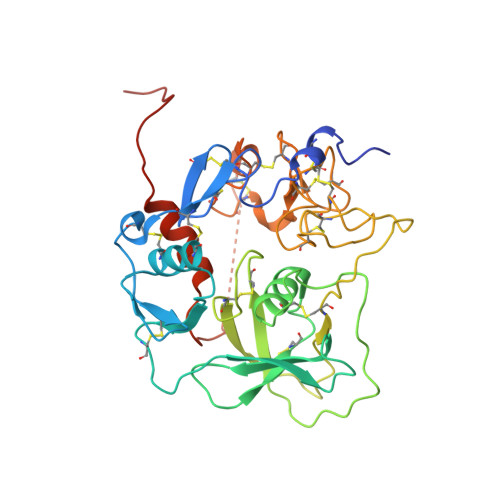
Regulator-dependent mechanisms of C3b processing by factor I allow differentiation of immune responses.
Xue, X., Wu, J., Ricklin, D., Forneris, F., Di Crescenzio, P., Schmidt, C.Q., Granneman, J., Sharp, T.H., Lambris, J.D., Gros, P.(2017) Nat Struct Mol Biol 24: 643-651
- PubMed: 28671664
- DOI: https://doi.org/10.1038/nsmb.3427
- Primary Citation of Related Structures:
5O32, 5O35 - PubMed Abstract:
The complement system labels microbes and host debris for clearance. Degradation of surface-bound C3b is pivotal to direct immune responses and protect host cells. How the serine protease factor I (FI), assisted by regulators, cleaves either two or three distant peptide bonds in the CUB domain of C3b remains unclear. We present a crystal structure of C3b in complex with FI and regulator factor H (FH; domains 1-4 with 19-20). FI binds C3b-FH between FH domains 2 and 3 and a reoriented C3b C-terminal domain and docks onto the first scissile bond, while stabilizing its catalytic domain for proteolytic activity. One cleavage in C3b does not affect its overall structure, whereas two cleavages unfold CUB and dislodge the thioester-containing domain (TED), affecting binding of regulators and thereby determining the number of cleavages. These data explain how FI generates late-stage opsonins iC3b or C3dg in a context-dependent manner, to react to foreign, danger or healthy self signals.
Organizational Affiliation:
Crystal and Structural Chemistry, Bijvoet Center for Biomolecular Research, Department of Chemistry, Faculty of Science, Utrecht University, Utrecht, the Netherlands.