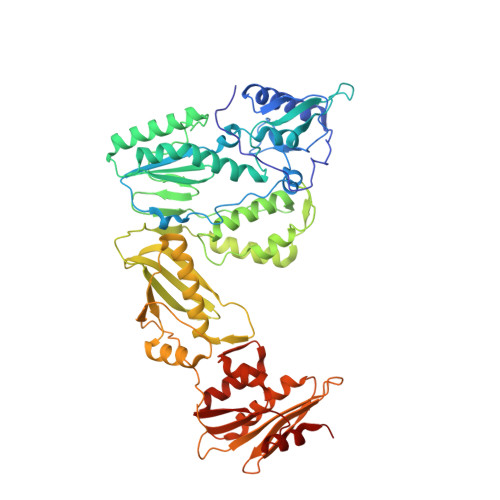
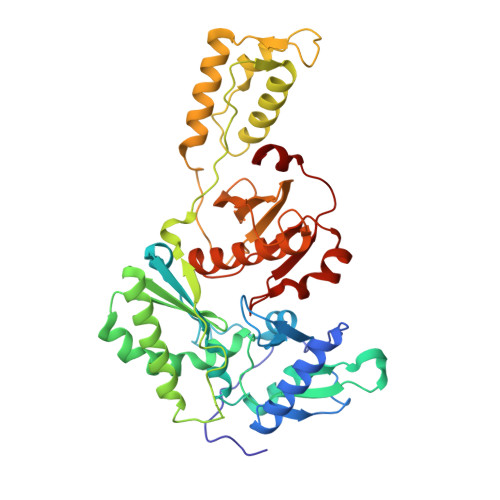
Design, synthesis and biological evaluations of N-Hydroxy thienopyrimidine-2,4-diones as inhibitors of HIV reverse transcriptase-associated RNase H.
Kankanala, J., Kirby, K.A., Huber, A.D., Casey, M.C., Wilson, D.J., Sarafianos, S.G., Wang, Z.(2017) Eur J Med Chem 141: 149-161
- PubMed: 29031062
- DOI: https://doi.org/10.1016/j.ejmech.2017.09.054
- Primary Citation of Related Structures:
6AOC - PubMed Abstract:
Human immunodeficiency virus (HIV) reverse transcriptase (RT) associated ribonuclease H (RNase H) is the only HIV enzymatic function not targeted by current antiviral drugs. Although various chemotypes have been reported to inhibit HIV RNase H, few have shown significant antiviral activities. We report herein the design, synthesis and biological evaluation of a novel N-hydroxy thienopyrimidine-2,3-dione chemotype (11) which potently and selectively inhibited RNase H with considerable potency against HIV-1 in cell culture. Current structure-activity-relationship (SAR) identified analogue 11d as a nanomolar inhibitor of RNase H (IC 50 = 0.04 μM) with decent antiviral potency (EC 50 = 7.4 μM) and no cytotoxicity (CC 50 > 100 μM). In extended biochemical assays compound 11d did not inhibit RT polymerase (pol) while inhibiting integrase strand transfer (INST) with 53 fold lower potency (IC 50 = 2.1 μM) than RNase H inhibition. Crystallographic and molecular modeling studies confirmed the RNase H active site binding mode.
- Center for Drug Design, Academic Health Center, University of Minnesota, Minneapolis, MN 55455, USA.
Organizational Affiliation: