Affinity maturation in a human humoral response to influenza hemagglutinin.
McCarthy, K.R., Raymond, D.D., Do, K.T., Schmidt, A.G., Harrison, S.C.(2019) Proc Natl Acad Sci U S A
- PubMed: 31843892
- DOI: https://doi.org/10.1073/pnas.1915620116
- Primary Citation of Related Structures:
6Q0E, 6Q0H, 6Q0I, 6Q0L, 6Q0O, 6Q18, 6Q19, 6Q1A, 6Q1E, 6Q1G, 6Q1J, 6Q1K - PubMed Abstract:
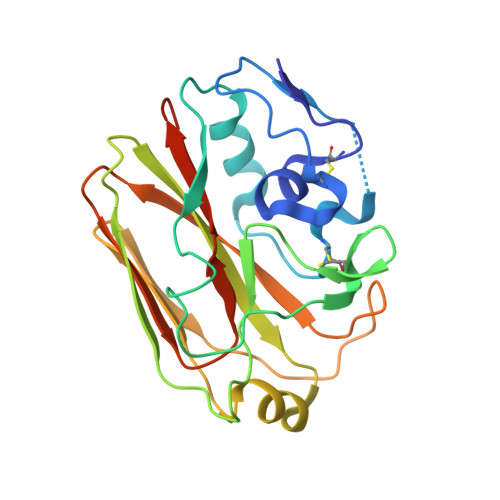
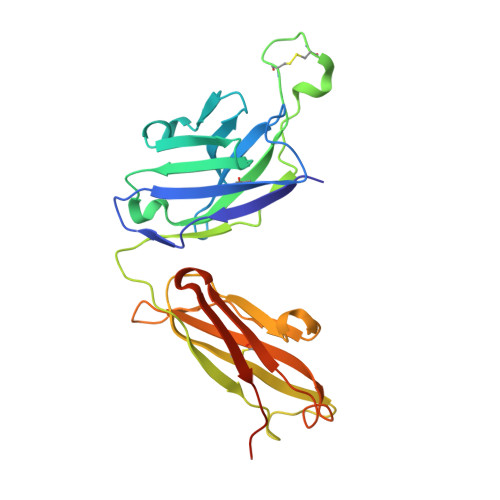
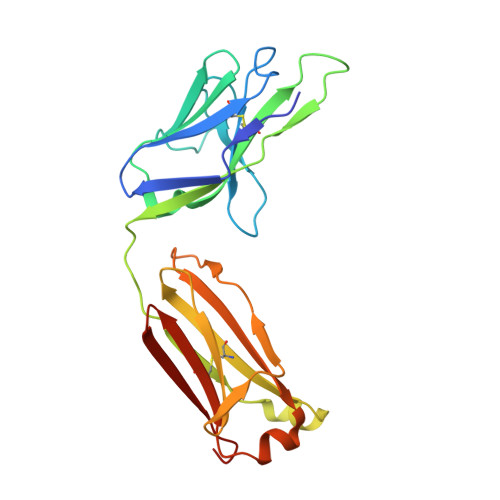
Affinity maturation of the B cell antigen receptor (BCR) is a conserved and crucial component of the adaptive immune response. BCR lineages, inferred from paired heavy- and light-chain sequences of rearranged Ig genes from multiple descendants of the same naive B cell precursor (the lineages' unmutated common ancestor, "UCA"), make it possible to reconstruct the underlying somatic evolutionary history. We present here an extensive structural and biophysical analysis of a lineage of BCRs directed against the receptor binding site (RBS) of subtype H1 influenza virus hemagglutinin (HA). The lineage includes 8 antibodies detected directly by sequencing, 3 in 1 principal branch and 5 in the other. When bound to HA, the heavy-chain third complementarity determining region (HCDR3) fits with an invariant pose into the RBS, but in each of the 2 branches, the rest of the Fab reorients specifically, from its position in the HA-bound UCA, about a hinge at the base of HCDR3. New contacts generated by the reorientation compensate for contacts lost as the H1 HA mutated during the time between the donor's initial exposure and the vaccination that preceded sampling. Our data indicate that a "pluripotent" naive response differentiated, in each branch, into 1 of its possible alternatives. This property of naive BCRs and persistence of multiple branches of their progeny lineages can offer broader protection from evolving pathogens than can a single, linear pathway of somatic mutation.
Organizational Affiliation:
Laboratory of Molecular Medicine, Boston Children's Hospital and Harvard Medical School, Boston, MA 02115.