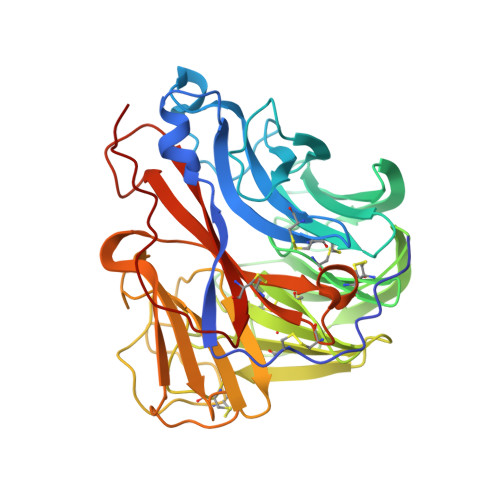
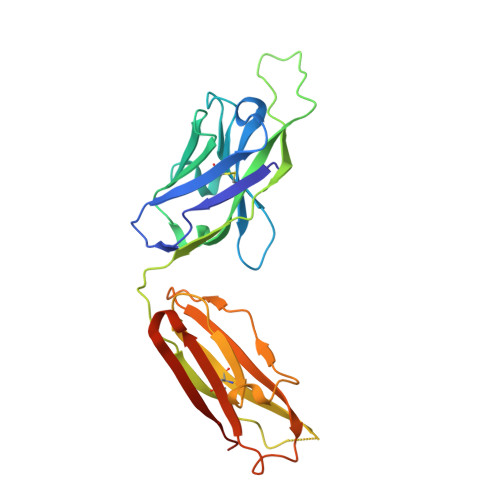
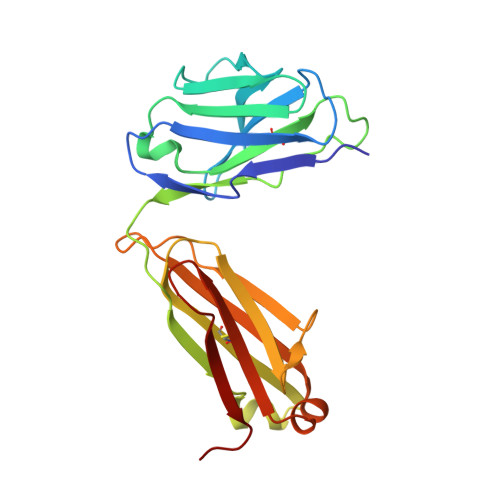
Broadly protective human antibodies that target the active site of influenza virus neuraminidase.
Stadlbauer, D., Zhu, X., McMahon, M., Turner, J.S., Wohlbold, T.J., Schmitz, A.J., Strohmeier, S., Yu, W., Nachbagauer, R., Mudd, P.A., Wilson, I.A., Ellebedy, A.H., Krammer, F.(2019) Science 366: 499-504
- PubMed: 31649200
- DOI: https://doi.org/10.1126/science.aay0678
- Primary Citation of Related Structures:
6Q1Z, 6Q20, 6Q23 - PubMed Abstract:
Better vaccines against influenza virus are urgently needed to provide broader protection against diverse strains, subtypes, and types. Such efforts are assisted by the identification of novel broadly neutralizing epitopes targeted by protective antibodies. Influenza vaccine development has largely focused on the hemagglutinin, but the other major surface antigen, the neuraminidase, has reemerged as a potential target for universal vaccines. We describe three human monoclonal antibodies isolated from an H3N2-infected donor that bind with exceptional breadth to multiple different influenza A and B virus neuraminidases. These antibodies neutralize the virus, mediate effector functions, are broadly protective in vivo, and inhibit neuraminidase activity by directly binding to the active site. Structural and functional characterization of these antibodies will inform the development of neuraminidase-based universal vaccines against influenza virus.
Organizational Affiliation:
Department of Microbiology, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA.