An Artificial Cofactor Catalyzing the Baylis-Hillman Reaction with Designed Streptavidin as Protein Host*.
Lechner, H., Emann, V.R., Breuning, M., Hocker, B.(2021) Chembiochem 22: 1573-1577
- PubMed: 33400831
- DOI: https://doi.org/10.1002/cbic.202000880
- Primary Citation of Related Structures:
6T1E, 6T1G, 6T1K, 6T2L, 6T2Y, 6T2Z, 6T30, 6T31, 6T32 - PubMed Abstract:
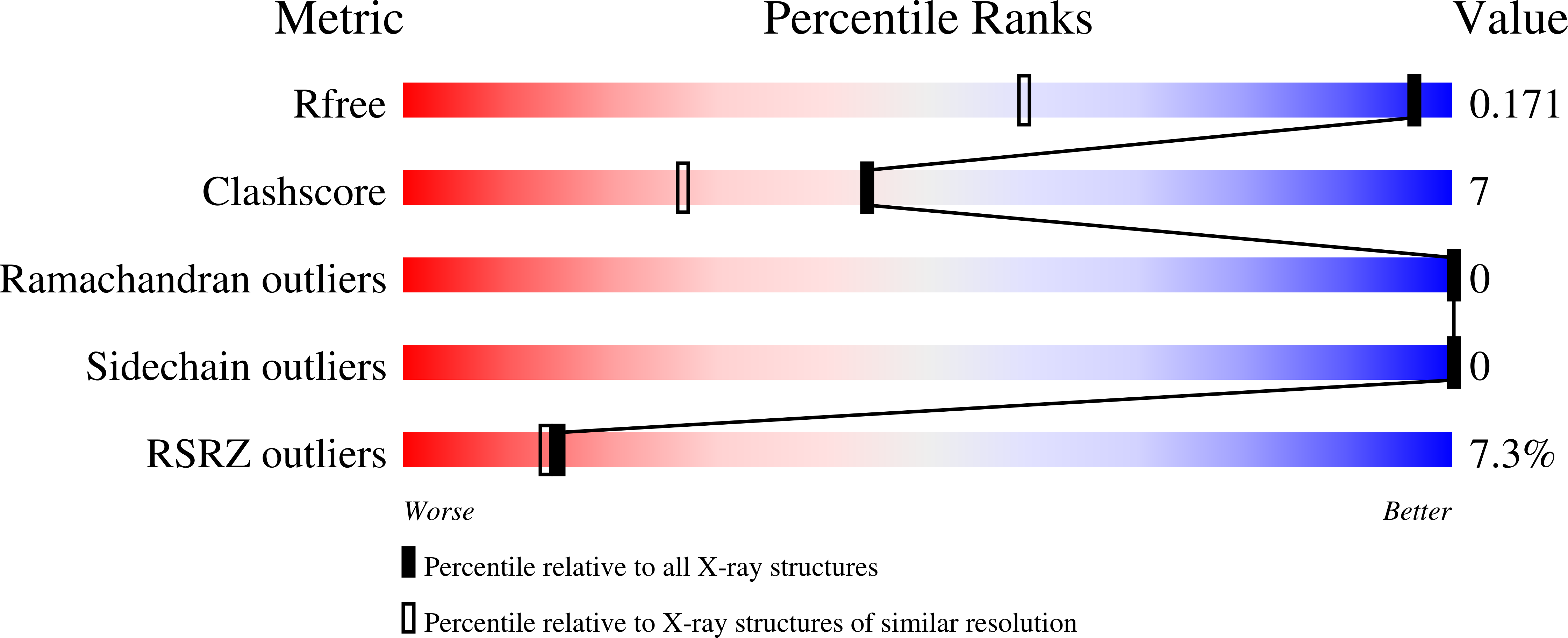
An artificial cofactor based on an organocatalyst embedded in a protein has been used to conduct the Baylis-Hillman reaction in a buffered system. As protein host, we chose streptavidin, as it can be easily crystallized and thereby supports the design process. The protein host around the cofactor was rationally designed on the basis of high-resolution crystal structures obtained after each variation of the amino acid sequence. Additionally, DFT-calculated intermediates and transition states were used to rationalize the observed activity. Finally, repeated cycles of structure determination and redesign led to a system with an up to one order of magnitude increase in activity over the bare cofactor and to the most active proteinogenic catalyst for the Baylis-Hillman reaction known today.
Organizational Affiliation:
Department of Biochemistry, University Bayreuth, Universitätsstrasse 30, 95447, Bayreuth, Germany.