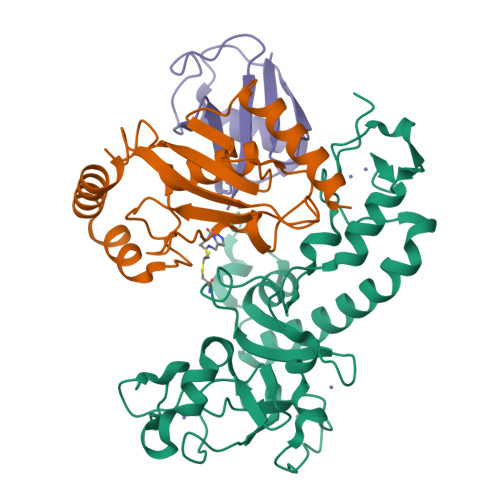
Structural basis for RING-Cys-Relay E3 ligase activity and its role in axon integrity.
Mabbitt, P.D., Loreto, A., Dery, M.A., Fletcher, A.J., Stanley, M., Pao, K.C., Wood, N.T., Coleman, M.P., Virdee, S.(2020) Nat Chem Biol 16: 1227-1236
- PubMed: 32747811
- DOI: https://doi.org/10.1038/s41589-020-0598-6
- Primary Citation of Related Structures:
6T7F - PubMed Abstract:
MYCBP2 is a ubiquitin (Ub) E3 ligase (E3) that is essential for neurodevelopment and regulates axon maintenance. MYCBP2 transfers Ub to nonlysine substrates via a newly discovered RING-Cys-Relay (RCR) mechanism, where Ub is relayed from an upstream cysteine to a downstream substrate esterification site. The molecular bases for E2-E3 Ub transfer and Ub relay are unknown. Whether these activities are linked to the neural phenotypes is also unclear. We describe the crystal structure of a covalently trapped E2~Ub:MYCBP2 transfer intermediate revealing key structural rearrangements upon E2-E3 Ub transfer and Ub relay. Our data suggest that transfer to the dynamic upstream cysteine, whilst mitigating lysine activity, requires a closed-like E2~Ub conjugate with tempered reactivity, and Ub relay is facilitated by a helix-coil transition. Furthermore, neurodevelopmental defects and delayed injury-induced degeneration in RCR-defective knock-in mice suggest its requirement, and that of substrate esterification activity, for normal neural development and programmed axon degeneration.
Organizational Affiliation:
MRC Protein Phosphorylation and Ubiquitylation Unit, University of Dundee, Dundee, UK.