Discovery and optimization of novel pyridines as highly potent and selective glycogen synthase kinase 3 inhibitors.
Ramurthy, S., Pfister, K.B., Boyce, R.S., Brown, S.P., Costales, A.Q., Desai, M.C., Fang, E., Levine, B.H., Ng, S.C., Nuss, J.M., Ring, D.B., Shafer, C.M., Shu, W., Subramanian, S., Wagman, A.S., Wang, H., Bussiere, D.E.(2020) Bioorg Med Chem Lett 30: 126930-126930
- PubMed: 31926786
- DOI: https://doi.org/10.1016/j.bmcl.2019.126930
- Primary Citation of Related Structures:
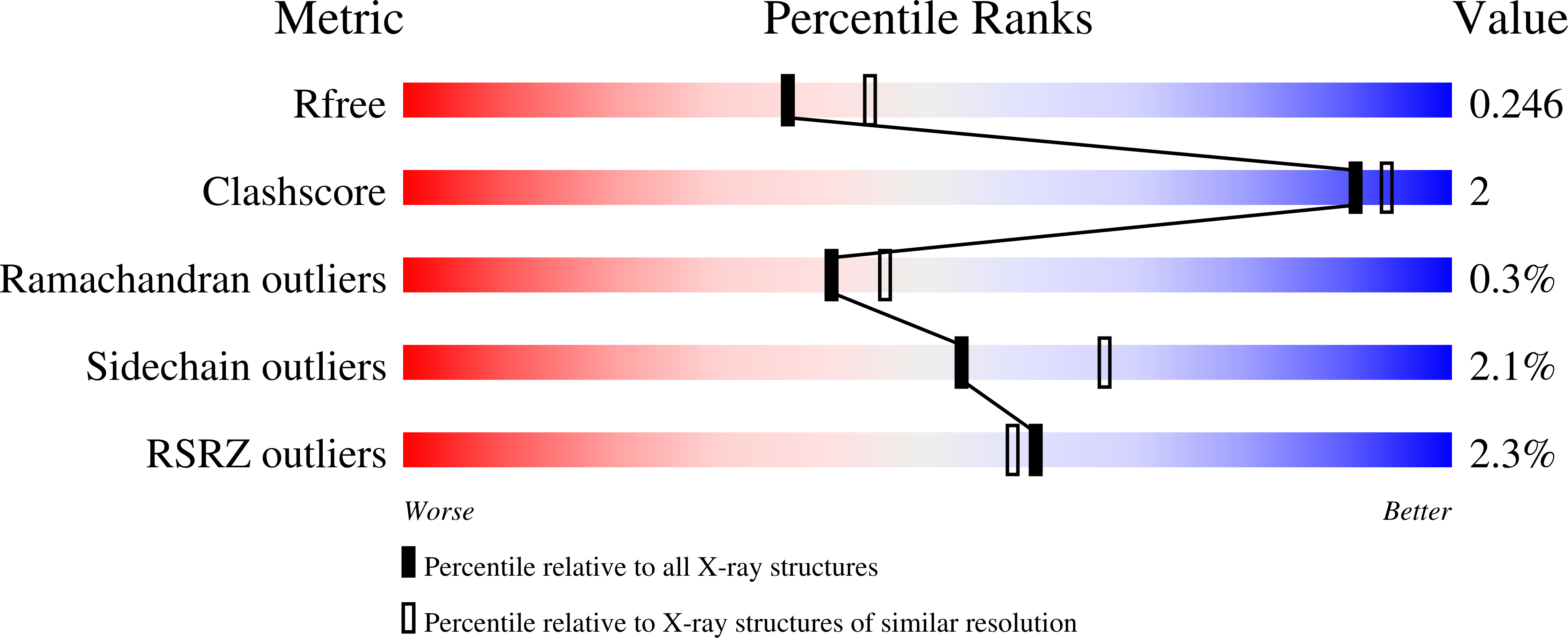
6V6L - PubMed Abstract:
Glycogen synthase kinase-3 plays an essential role in multiple biochemical pathways in the cell, particularly in regards to energy regulation. As such, Glycogen synthase kinase-3 is an attractive target for pharmacological intervention in a variety of disease states, particularly non-insulin dependent diabetes mellitus. However, due to homology with other crucial kinases, such as the cyclin-dependent protein kinase CDC2, developing compounds that are both potent and selective is challenging. A novel series of derivatives of 5-nitro-N2-(2-(pyridine-2ylamino)ethyl)pyridine-2,6-diamine were synthesized and have been shown to potently inhibit glycogen synthase kinase-3 (GSK3). Potency in the low nanomolar range was obtained along with remarkable selectivity. The compounds activate glycogen synthase in insulin receptor-expressing CHO-IR cells and in primary rat hepatocytes, and have acceptable pharmacokinetics and pharmacodynamics to allow for oral dosing. The X-ray co-crystal structure of human GSK3-β in complex with compound 2 is reported and provides insights into the structural determinants of the series responsible for its potency and selectivity.
Organizational Affiliation:
HiberCell, Inc., 619 West 54th Street, New York, NY 10019, USA.