Low-dose in situ prelocation of protein microcrystals by 2D X-ray phase-contrast imaging for serial crystallography.
Martiel, I., Huang, C.Y., Villanueva-Perez, P., Panepucci, E., Basu, S., Caffrey, M., Pedrini, B., Bunk, O., Stampanoni, M., Wang, M.(2020) IUCrJ 7: 1131-1141
- PubMed: 33209324
- DOI: https://doi.org/10.1107/S2052252520013238
- Primary Citation of Related Structures:
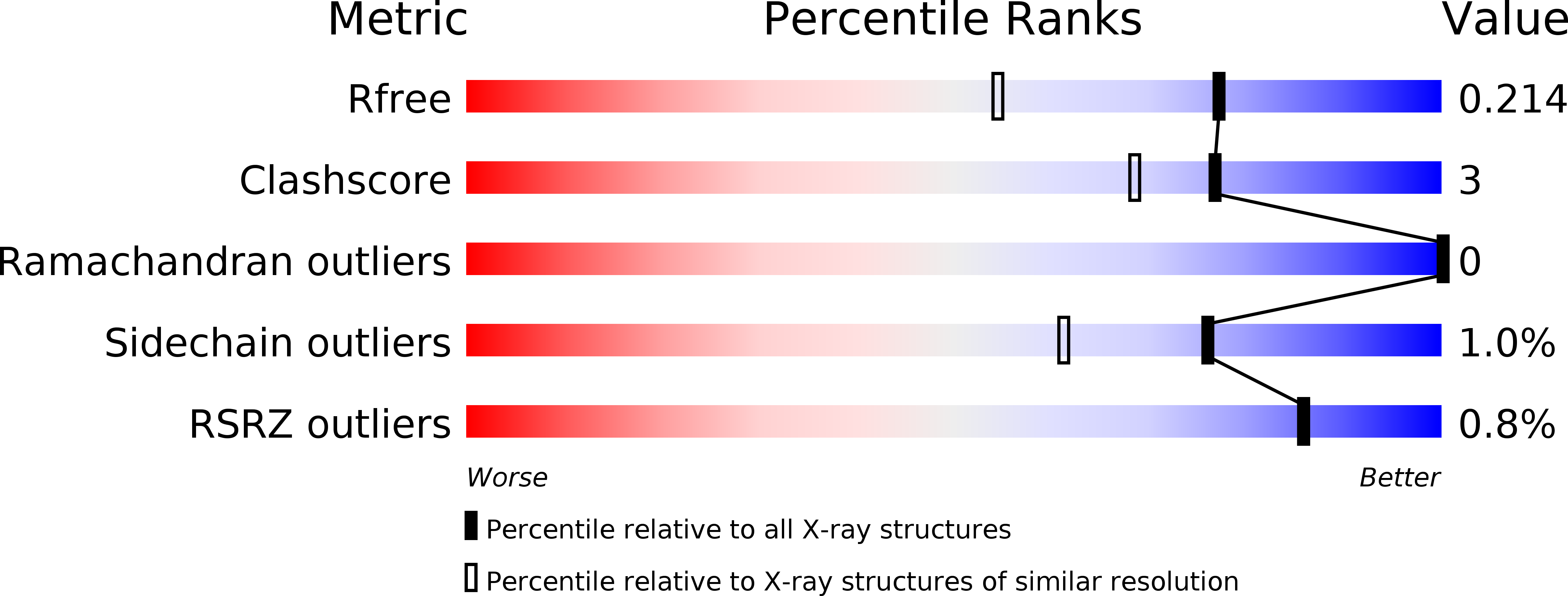
6YOB, 6YOC, 6YOD, 6YOE, 6YOF, 6YOG - PubMed Abstract:
Serial protein crystallography has emerged as a powerful method of data collection on small crystals from challenging targets, such as membrane proteins. Multiple microcrystals need to be located on large and often flat mounts while exposing them to an X-ray dose that is as low as possible. A crystal-prelocation method is demonstrated here using low-dose 2D full-field propagation-based X-ray phase-contrast imaging at the X-ray imaging beamline TOMCAT at the Swiss Light Source (SLS). This imaging step provides microcrystal coordinates for automated serial data collection at a microfocus macromolecular crystallography beamline on samples with an essentially flat geometry. This prelocation method was applied to microcrystals of a soluble protein and a membrane protein, grown in a commonly used double-sandwich in situ crystallization plate. The inner sandwiches of thin plastic film enclosing the microcrystals in lipid cubic phase were flash cooled and imaged at TOMCAT. Based on the obtained crystal coordinates, both still and rotation wedge serial data were collected automatically at the SLS PXI beamline, yielding in both cases a high indexing rate. This workflow can be easily implemented at many synchrotron facilities using existing equipment, or potentially integrated as an online technique in the next-generation macromolecular crystallography beamline, and thus benefit a number of dose-sensitive challenging protein targets.
Organizational Affiliation:
Paul Scherrer Institute, Forschungsstrasse 111, Villigen, 5232, Switzerland.