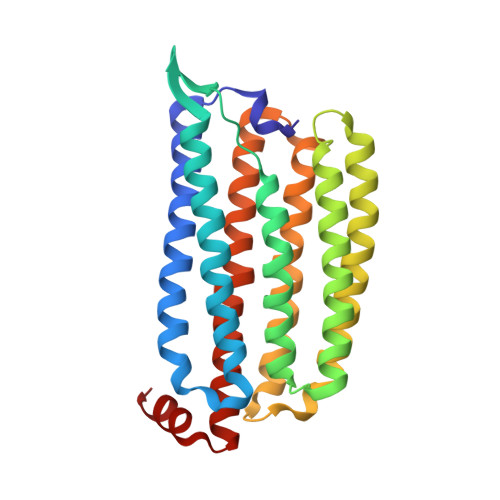
Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Yun, J.H., Li, X., Yue, J., Park, J.H., Jin, Z., Li, C., Hu, H., Shi, Y., Pandey, S., Carbajo, S., Boutet, S., Hunter, M.S., Liang, M., Sierra, R.G., Lane, T.J., Zhou, L., Weierstall, U., Zatsepin, N.A., Ohki, M., Tame, J.R.H., Park, S.Y., Spence, J.C.H., Zhang, W., Schmidt, M., Lee, W., Liu, H.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 33753488
- DOI: https://doi.org/10.1073/pnas.2020486118
- Primary Citation of Related Structures:
7CRI, 7CRJ, 7CRK, 7CRL, 7CRS, 7CRT, 7CRX, 7CRY - PubMed Abstract:
Chloride ion-pumping rhodopsin (ClR) in some marine bacteria utilizes light energy to actively transport Cl - into cells. How the ClR initiates the transport is elusive. Here, we show the dynamics of ion transport observed with time-resolved serial femtosecond (fs) crystallography using the Linac Coherent Light Source. X-ray pulses captured structural changes in ClR upon flash illumination with a 550 nm fs-pumping laser. High-resolution structures for five time points (dark to 100 ps after flashing) reveal complex and coordinated dynamics comprising retinal isomerization, water molecule rearrangement, and conformational changes of various residues. Combining data from time-resolved spectroscopy experiments and molecular dynamics simulations, this study reveals that the chloride ion close to the Schiff base undergoes a dissociation-diffusion process upon light-triggered retinal isomerization.
Organizational Affiliation:
Department of Biochemistry, College of Life Sciences and Biotechnology, Yonsei University, Seodaemun-gu, 120-749 Seoul, South Korea.