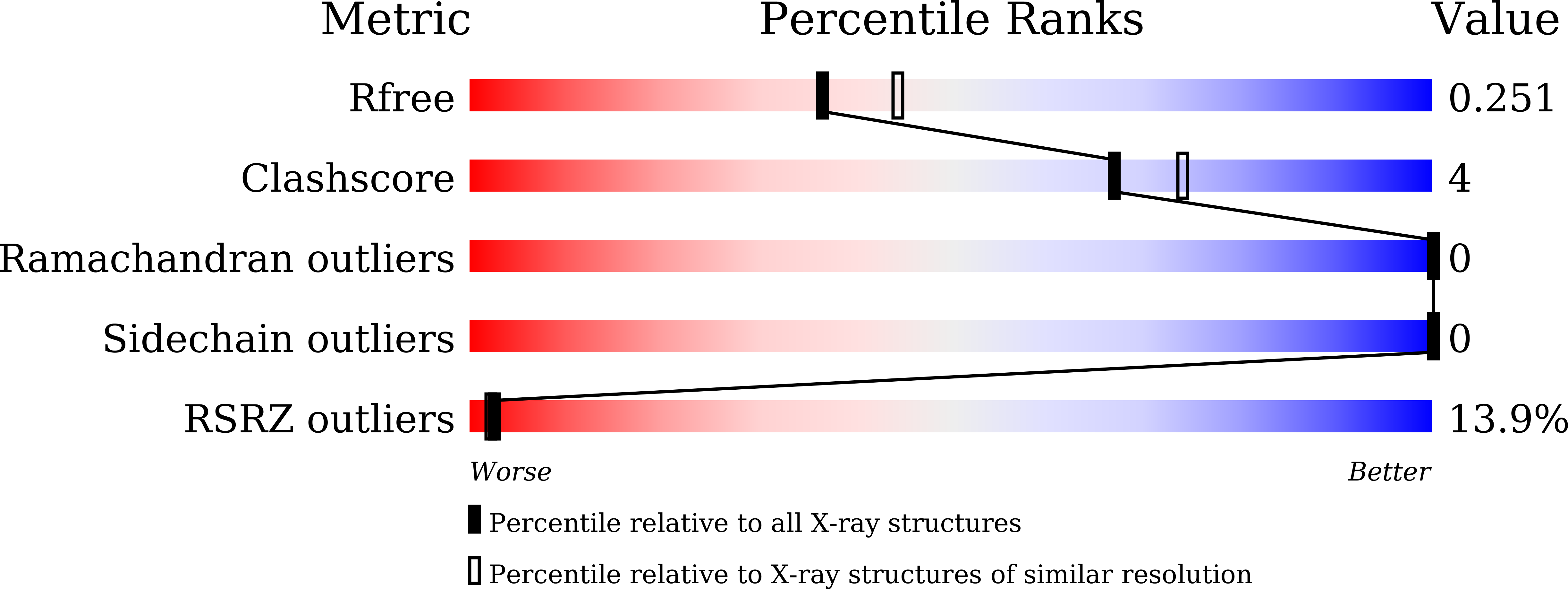
Investigate Natural Product Indolmycin and the Synthetically Improved Analogue Toward Antimycobacterial Agents.
Yang, Y., Xu, Y., Yue, Y., Wang, H., Cui, Y., Pan, M., Zhang, X., Zhang, L., Li, H., Xu, M., Tang, Y., Chen, S.(2022) ACS Chem Biol 17: 39-53
- PubMed: 34908399
- DOI: https://doi.org/10.1021/acschembio.1c00394
- Primary Citation of Related Structures:
7EL8, 7ELT, 7ENS, 7ENT, 7EV2, 7EV3 - PubMed Abstract:
Indolmycin (IND) is a microbial natural product that selectively inhibits bacterial tryptophanyl-tRNA synthetase (TrpRS). The tryptophan biosynthesis pathway was recently shown to be an important target for developing new antibacterial agents against Mycobacterium tuberculosis (Mtb). We investigated the antibacterial activity of IND against several mycobacterial model strains. A TrpRS biochemical assay was developed to analyze a library of synthetic IND analogues. The 4″-methylated IND compound, Y-13, showed improved anti-Mtb activity with a minimum inhibitory concentration (MIC) of 1.88 μM (∼0.5 μg/mL). The MIC increased significantly when overexpression of TrpRS was induced in the genetically engineered surrogate M. bovis BCG. The cocrystal structure of Mtb TrpRS complexed with IND and ATP has revealed that the amino acid pocket is in a state between the open form of apo protein and the closed complex with the reaction intermediate. In whole-cell-based experiments, we studied the combination effect of Y-13 paired with different antibacterial agents. We evaluated the killing kinetics, the frequency of resistance to INDs, and the mode of resistance of IND-resistant mycobacteria by genome sequencing. The synergistic interaction of Y-13 with the TrpE allosteric inhibitor, indole propionic acid, suggests that prospective IND analogues could shut down tryptophan biosynthesis and protein biosynthesis in pathogens, leading to a new class of antibiotics. Finally, we discuss a strategy to expand the genome mining of antibiotic-producing microbes specifically for antimycobacterial development.
Organizational Affiliation:
School of Pharmaceutical Sciences, MOE Key Laboratory of Bioorganic Phosphorus Chemistry & Chemical Biology, Tsinghua University, Beijing 100084, China.