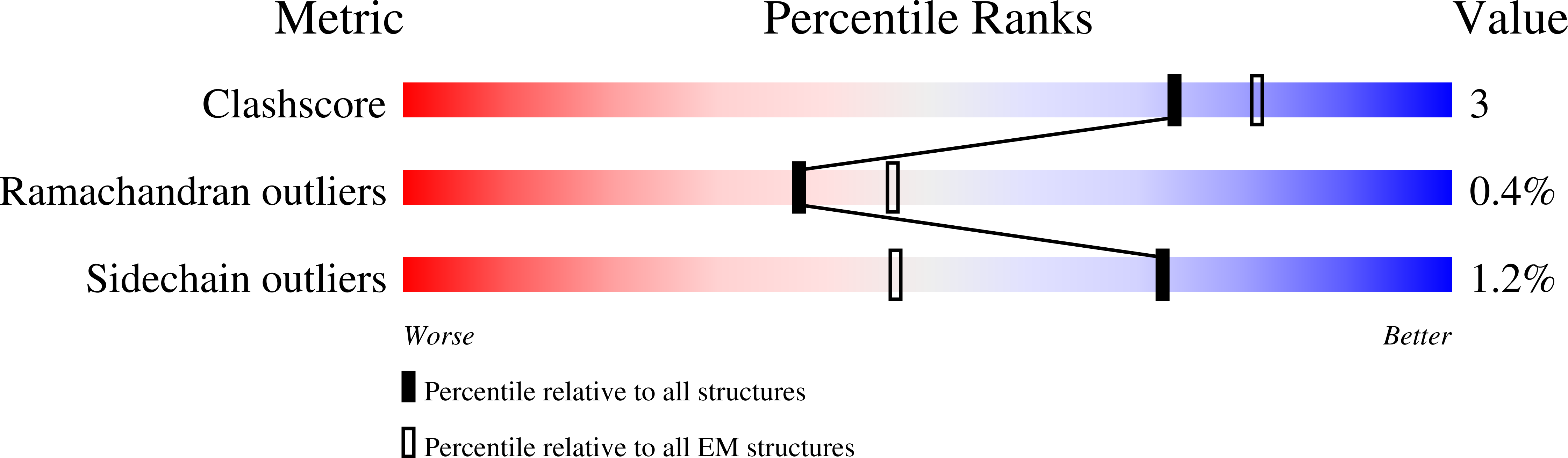IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Burrell, A.L., Nie, C., Said, M., Simonet, J.C., Fernandez-Justel, D., Johnson, M.C., Quispe, J., Buey, R.M., Peterson, J.R., Kollman, J.M.(2022) Nat Struct Mol Biol 29: 47-58
- PubMed: 35013599
- DOI: https://doi.org/10.1038/s41594-021-00706-2
- Primary Citation of Related Structures:
7RER, 7RES, 7RFE, 7RFF, 7RFG, 7RFH, 7RFI, 7RGD, 7RGI, 7RGL, 7RGM, 7RGQ - PubMed Abstract:
Inosine-5'-monophosphate dehydrogenase (IMPDH), a key regulatory enzyme in purine nucleotide biosynthesis, dynamically assembles filaments in response to changes in metabolic demand. Humans have two isoforms: IMPDH2 filaments reduce sensitivity to feedback inhibition, while IMPDH1 assembly remains uncharacterized. IMPDH1 plays a unique role in retinal metabolism, and point mutants cause blindness. Here, in a series of cryogenic-electron microscopy structures we show that human IMPDH1 assembles polymorphic filaments with different assembly interfaces in extended and compressed states. Retina-specific splice variants introduce structural elements that reduce sensitivity to GTP inhibition, including stabilization of the extended filament form. Finally, we show that IMPDH1 disease mutations fall into two classes: one disrupts GTP regulation and the other has no effect on GTP regulation or filament assembly. These findings provide a foundation for understanding the role of IMPDH1 in retinal function and disease and demonstrate the diverse mechanisms by which metabolic enzyme filaments are allosterically regulated.
Organizational Affiliation:
Department of Biochemistry, University of Washington, Seattle, WA, USA.