Crystal structure of delta sub IV Rhodobacter Sphaeroides bc1 with the antimalarial drug atovaquone.
Esser, L., Xia, D., Zhou, F.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome b | A, D [auth E], G [auth K], J [auth O] | 445 | Cereibacter sphaeroides | Mutation(s): 0 Gene Names: petB, fbcB Membrane Entity: Yes | 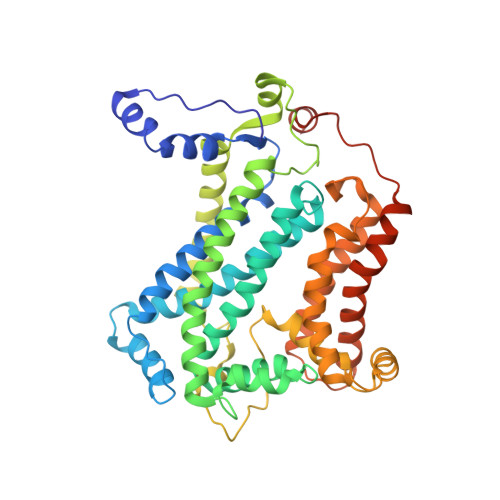 |
UniProt | |||||
Find proteins for Q02761 (Cereibacter sphaeroides) Explore Q02761 Go to UniProtKB: Q02761 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q02761 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c1 | B, E [auth F], H [auth L], K [auth P] | 269 | Cereibacter sphaeroides | Mutation(s): 0 Membrane Entity: Yes | 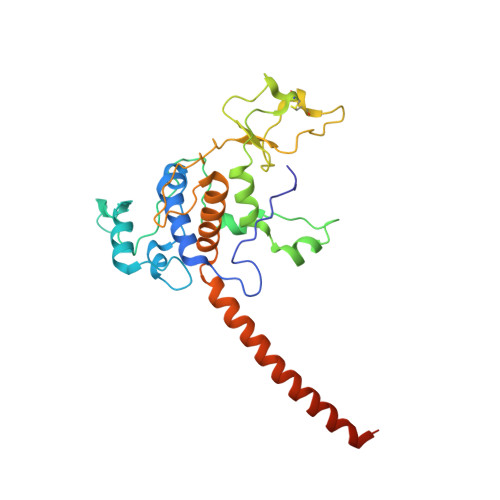 |
UniProt | |||||
Find proteins for Q02760 (Cereibacter sphaeroides) Explore Q02760 Go to UniProtKB: Q02760 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q02760 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ubiquinol-cytochrome c reductase iron-sulfur subunit | C, F [auth G], I [auth M], L [auth Q] | 187 | Cereibacter sphaeroides | Mutation(s): 0 Gene Names: petA, fbcF EC: 7.1.1.8 Membrane Entity: Yes | 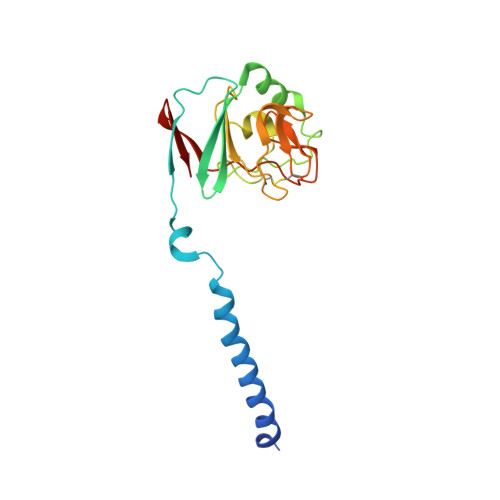 |
UniProt | |||||
Find proteins for Q02762 (Cereibacter sphaeroides) Explore Q02762 Go to UniProtKB: Q02762 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q02762 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 7 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| HEC Query on HEC | BA [auth F], JA [auth L], RA [auth P], S [auth B] | HEME C C34 H34 Fe N4 O4 HXQIYSLZKNYNMH-LJNAALQVSA-N |  | ||
| HEM Query on HEM | EA [auth K] FA [auth K] M [auth A] MA [auth O] N [auth A] | PROTOPORPHYRIN IX CONTAINING FE C34 H32 Fe N4 O4 KABFMIBPWCXCRK-RGGAHWMASA-L |  | ||
| 6PE Query on 6PE | HA [auth K], P [auth A], PA [auth O], Y [auth E] | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE C17 H33 N O8 P PELYUHWUVHDSSU-OAHLLOKOSA-M |  | ||
| AOQ (Subject of Investigation/LOI) Query on AOQ | GA [auth K], O [auth A], OA [auth O], X [auth E] | 2-[trans-4-(4-chlorophenyl)cyclohexyl]-3-hydroxynaphthalene-1,4-dione C22 H19 Cl O3 KUCQYCKVKVOKAY-CTYIDZIISA-N |  | ||
| BOG Query on BOG | IA [auth K], QA [auth O], R [auth A], Z [auth E] | octyl beta-D-glucopyranoside C14 H28 O6 HEGSGKPQLMEBJL-RKQHYHRCSA-N |  | ||
| FES Query on FES | DA [auth G], LA [auth M], TA [auth Q], U [auth C] | FE2/S2 (INORGANIC) CLUSTER Fe2 S2 NIXDOXVAJZFRNF-UHFFFAOYSA-N |  | ||
| SR Query on SR | AA [auth E] CA [auth F] KA [auth L] Q [auth A] SA [auth P] | STRONTIUM ION Sr PWYYWQHXAPXYMF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 127.273 | α = 90 |
| b = 156.388 | β = 96.67 |
| c = 141.407 | γ = 90 |
| Software Name | Purpose |
|---|---|
| SCALEPACK | data scaling |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data reduction |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Cancer Institute (NIH/NCI) | United States | -- |