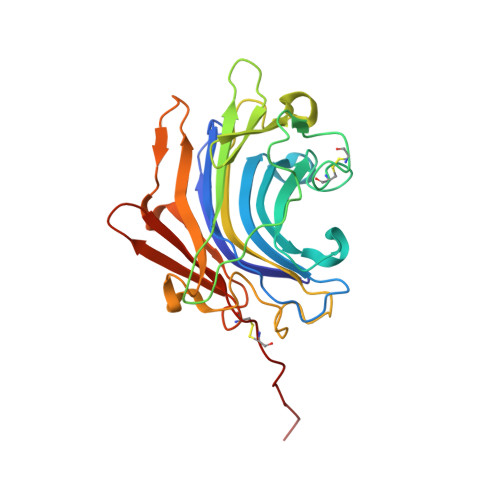
Molecular and in vivo studies of a glutamate-class prolyl-endopeptidase for coeliac disease therapy.
Del Amo-Maestro, L., Mendes, S.R., Rodriguez-Banqueri, A., Garzon-Flores, L., Girbal, M., Rodriguez-Lagunas, M.J., Guevara, T., Franch, A., Perez-Cano, F.J., Eckhard, U., Gomis-Ruth, F.X.(2022) Nat Commun 13: 4446-4446
- PubMed: 35915115
- DOI: https://doi.org/10.1038/s41467-022-32215-1
- Primary Citation of Related Structures:
7ZU8, 7ZVA, 7ZVB, 7ZVC - PubMed Abstract:
The digestion of gluten generates toxic peptides, among which a highly immunogenic proline-rich 33-mer from wheat α-gliadin, that trigger coeliac disease. Neprosin from the pitcher plant is a reported prolyl endopeptidase. Here, we produce recombinant neprosin and its mutants, and find that full-length neprosin is a zymogen, which is self-activated at gastric pH by the release of an all-β pro-domain via a pH-switch mechanism featuring a lysine plug. The catalytic domain is an atypical 7+8-stranded β-sandwich with an extended active-site cleft containing an unprecedented pair of catalytic glutamates. Neprosin efficiently degrades both gliadin and the 33-mer in vitro under gastric conditions and is reversibly inactivated at pH > 5. Moreover, co-administration of gliadin and the neprosin zymogen at the ratio 500:1 reduces the abundance of the 33-mer in the small intestine of mice by up to 90%. Neprosin therefore founds a family of eukaryotic glutamate endopeptidases that fulfils requisites for a therapeutic glutenase.
- Proteolysis Laboratory; Department of Structural Biology, Molecular Biology Institute of Barcelona (CSIC), Barcelona Science Park; c/Baldiri Reixac, 15-21, 08028, Barcelona, Catalonia, Spain.
Organizational Affiliation: