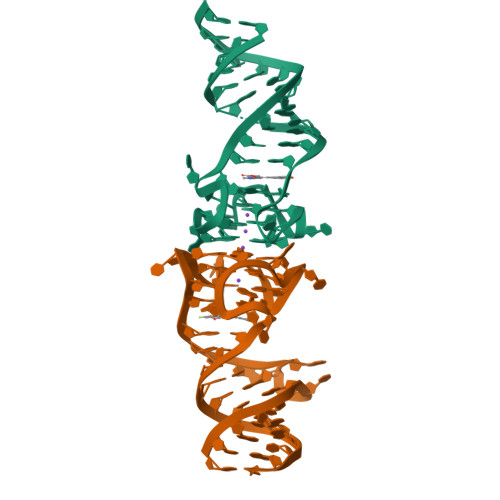
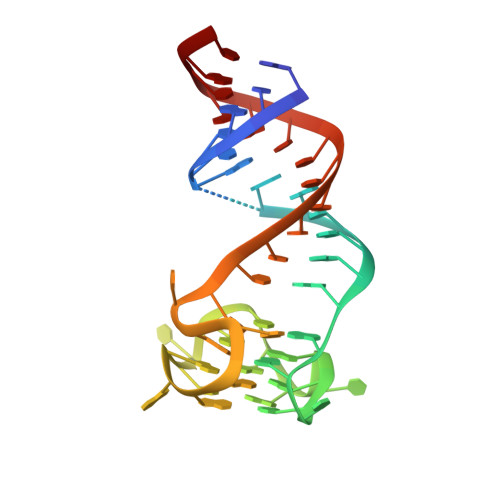
Co-crystal structures of the fluorogenic aptamer Beetroot show that close homology may not predict similar RNA architecture.
Passalacqua, L.F.M., Starich, M.R., Link, K.A., Wu, J., Knutson, J.R., Tjandra, N., Jaffrey, S.R., Ferre-D'Amare, A.R.(2023) Nat Commun 14: 2969-2969
- PubMed: 37221204
- DOI: https://doi.org/10.1038/s41467-023-38683-3
- Primary Citation of Related Structures:
8EYU, 8EYV, 8EYW, 8F0N - PubMed Abstract:
Beetroot is a homodimeric in vitro selected RNA that binds and activates DFAME, a conditional fluorophore derived from GFP. It is 70% sequence-identical to the previously characterized homodimeric aptamer Corn, which binds one molecule of its cognate fluorophore DFHO at its interprotomer interface. We have now determined the Beetroot-DFAME co-crystal structure at 1.95 Å resolution, discovering that this RNA homodimer binds two molecules of the fluorophore, at sites separated by ~30 Å. In addition to this overall architectural difference, the local structures of the non-canonical, complex quadruplex cores of Beetroot and Corn are distinctly different, underscoring how subtle RNA sequence differences can give rise to unexpected structural divergence. Through structure-guided engineering, we generated a variant that has a 12-fold fluorescence activation selectivity switch toward DFHO. Beetroot and this variant form heterodimers and constitute the starting point for engineered tags whose through-space inter-fluorophore interaction could be used to monitor RNA dimerization.
Organizational Affiliation:
Biochemistry and Biophysics Center, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD, USA.