Structural analysis of G protein inhibition by cyclic depsipeptide inhibitors
Muehle, J., Schertler, G.F.X.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein subunit alpha-11 | 352 | Homo sapiens | Mutation(s): 0 Gene Names: GNA11, GA11 EC: 3.6.5 | 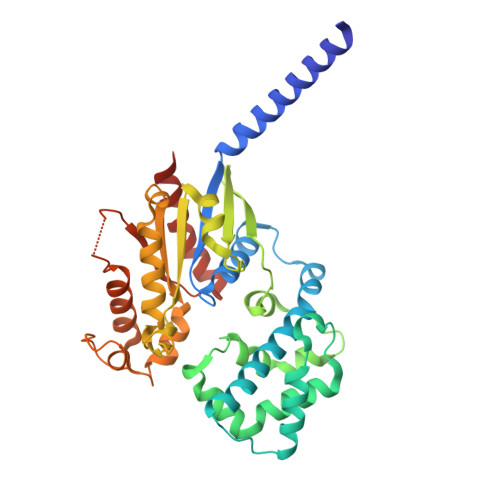 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P29992 (Homo sapiens) Explore P29992 Go to UniProtKB: P29992 | |||||
PHAROS: P29992 GTEx: ENSG00000088256 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P29992 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | 344 | Homo sapiens | Mutation(s): 0 Gene Names: GNB1 | 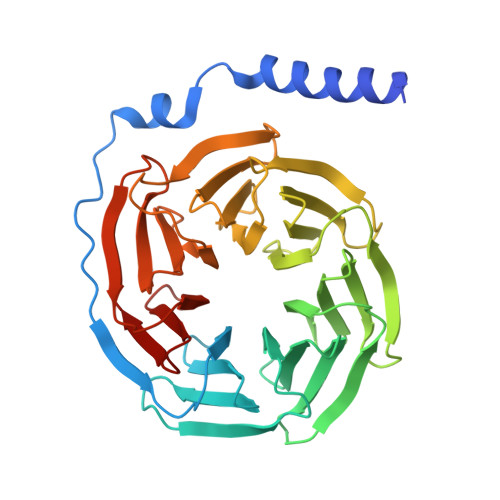 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P62873 (Homo sapiens) Explore P62873 Go to UniProtKB: P62873 | |||||
PHAROS: P62873 GTEx: ENSG00000078369 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62873 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 | C [auth G] | 71 | Homo sapiens | Mutation(s): 1 Gene Names: GNG2 | 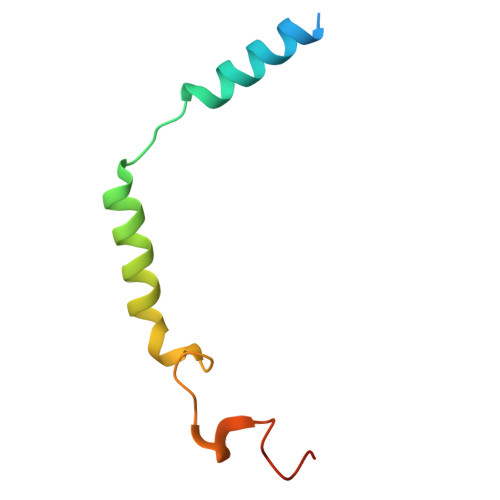 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P59768 (Homo sapiens) Explore P59768 Go to UniProtKB: P59768 | |||||
PHAROS: P59768 GTEx: ENSG00000186469 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P59768 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 10 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GDP Query on GDP | D [auth A] | GUANOSINE-5'-DIPHOSPHATE C10 H15 N5 O11 P2 QGWNDRXFNXRZMB-UUOKFMHZSA-N |  | ||
| HF2 (Subject of Investigation/LOI) Query on HF2 | K [auth A] | (2R)-2-hydroxy-3-phenylpropanoic acid C9 H10 O3 VOXXWSYKYCBWHO-MRVPVSSYSA-N |  | ||
| THC (Subject of Investigation/LOI) Query on THC | L [auth A] | N-METHYLCARBONYLTHREONINE C6 H11 N O4 PEDXUVCGOLSNLQ-WUJLRWPWSA-N |  | ||
| HL2 (Subject of Investigation/LOI) Query on HL2 | N [auth A], O [auth A] | (2S,3R)-2-amino-3-hydroxy-4-methylpentanoic acid C6 H13 N O3 ZAYJDMWJYCTABM-CRCLSJGQSA-N |  | ||
| OTH (Subject of Investigation/LOI) Query on OTH | M [auth A] | N,O-dimethyl-L-threonine C6 H13 N O3 ZLRWZUVKLXZLRT-UHNVWZDZSA-N |  | ||
| MAA (Subject of Investigation/LOI) Query on MAA | S [auth B] | N-methyl-L-alanine C4 H9 N O2 GDFAOVXKHJXLEI-VKHMYHEASA-N |  | ||
| DAM (Subject of Investigation/LOI) Query on DAM | J [auth A] | N-METHYL-ALPHA-BETA-DEHYDROALANINE C4 H7 N O2 FLEYLGCAQDCGHN-UHFFFAOYSA-N |  | ||
| ALA (Subject of Investigation/LOI) Query on ALA | T [auth B] | ALANINE C3 H7 N O2 QNAYBMKLOCPYGJ-REOHCLBHSA-N |  | ||
| EDO Query on EDO | E [auth A] F [auth A] G [auth A] H [auth A] I [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| ACE (Subject of Investigation/LOI) Query on ACE | U [auth B] | ACETYL GROUP C2 H4 O IKHGUXGNUITLKF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 72.604 | α = 90 |
| b = 102.533 | β = 90 |
| c = 154.002 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| autoPROC | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| German Research Foundation (DFG) | Germany | 273251628 |