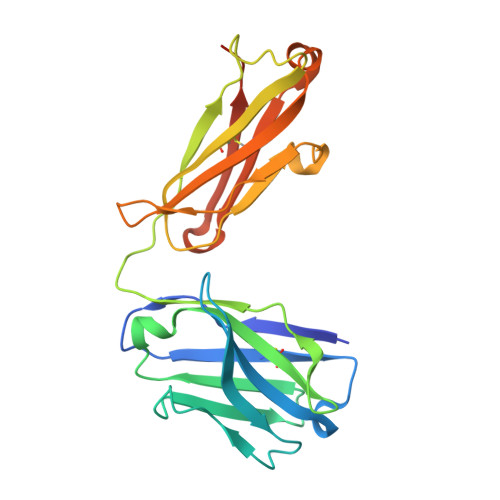
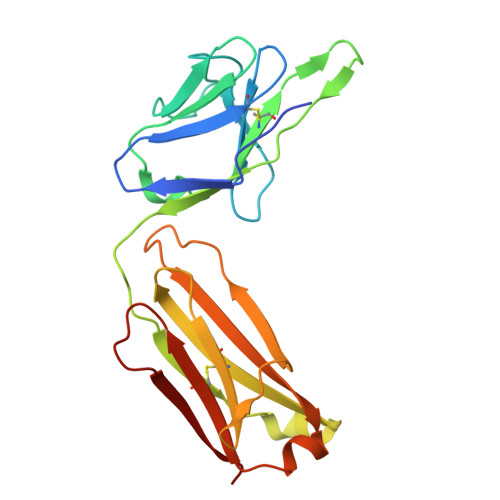
Crystal structure of an antibody specifically recognizing 3,4-methyl enedioxy methamphetamine through the epoxide moiety.
Cheon, G., Hwang, D., Le, T.C., Lee, Y., Han, E., An, S., Jung, Y., Chung, H., Lee, S.(2024) Biochem Biophys Res Commun 733: 150607-150607
- PubMed: 39208641
- DOI: https://doi.org/10.1016/j.bbrc.2024.150607
- Primary Citation of Related Structures:
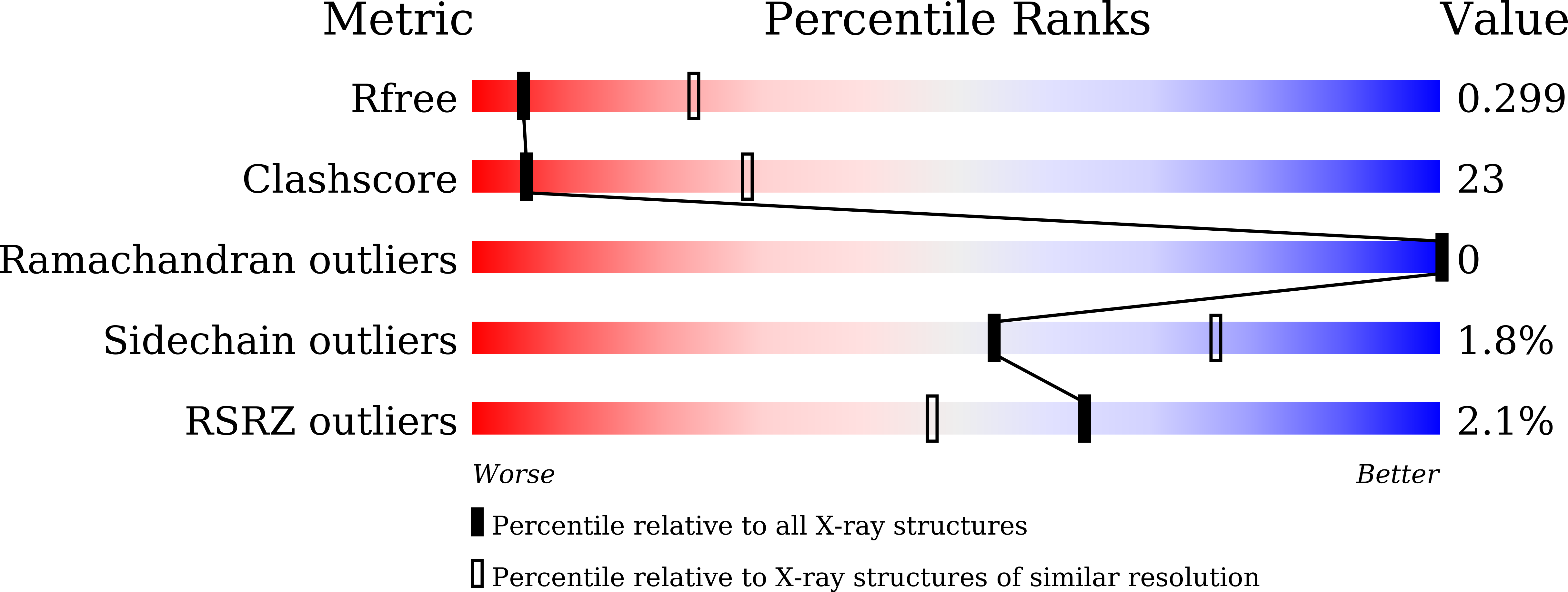
8Y57 - PubMed Abstract:
3,4-methylenedioxymethamphetamine (MDMA) or publicly known as "ecstasy" is a drug abuse substance. Since antibodies that detect MDMA typically also recognize its chemical analogue, methamphetamine (METH), we identified antibodies specifically recognizing MDMA, but not METH, named 1bB11 and 1bF12, using phage display. The crystal structure of 1bB11 in complex with MDMA was determined at 3.2 Å resolution. Key interactions were found between the epoxide moiety of MDMA and S34 and Y36 of the light chain. Additional interaction with E33 of the heavy chain contributes to anchoring MDMA. Mutagenesis-based biochemical analysis confirmed the importance of these residues in MDMA binding. Comparing the structure of 1bB11 to a scFv6H4, which binds both METH and MDMA, revealed opposite binding orientations. Taken together, our data provides a structural framework for selective binding to MDMA by the 1bB11 antibody, paving a way to develop a highly specific antibody for diagnosis.
Organizational Affiliation:
Department of Biological Sciences, Sungkyunkwan University, Suwon, 16419, Republic of Korea.