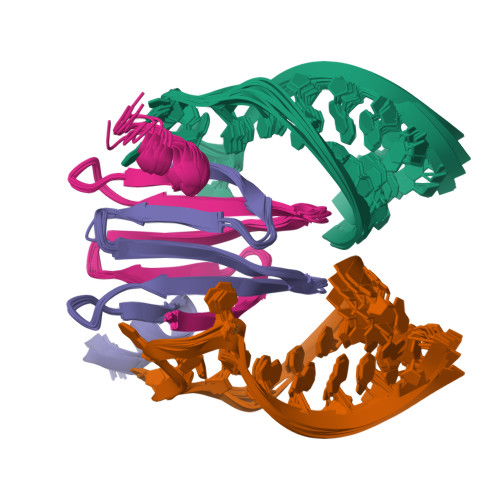
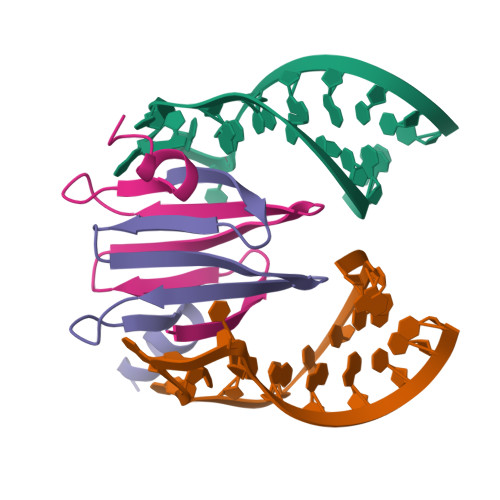
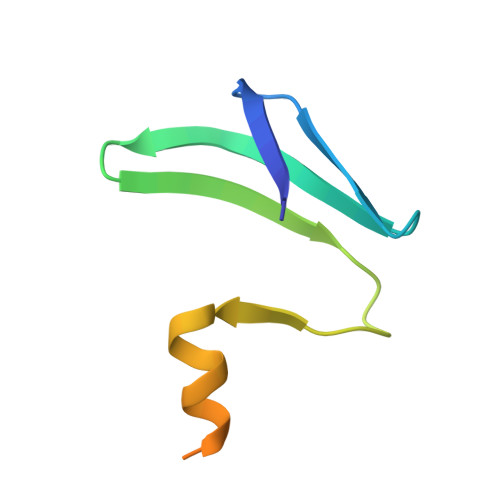
Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA
Schubert, M., Lapouge, K., Duss, O., Oberstrass, F.C., Jelesarov, I., Haas, D., Allain, F.H.-T.(2007) Nat Struct Mol Biol 14: 807-813
- PubMed: 17704818
- DOI: https://doi.org/10.1038/nsmb1285
- Primary Citation of Related Structures:
2JPP - PubMed Abstract:
Proteins of the RsmA/CsrA family are global translational regulators in many bacterial species. We have determined the solution structure of a complex formed between the RsmE protein, a member of this family from Pseudomonas fluorescens, and a target RNA encompassing the ribosome-binding site of the hcnA gene. The RsmE homodimer with its two RNA-binding sites makes optimal contact with an 5'-A/UCANGGANGU/A-3' sequence in the mRNA. When tightly gripped by RsmE, the ANGGAN core folds into a loop, favoring the formation of a 3-base-pair stem by flanking nucleotides. We validated these findings by in vivo and in vitro mutational analyses. The structure of the complex explains well how, by sequestering the Shine-Dalgarno sequence, the RsmA/CsrA proteins repress translation.
Organizational Affiliation:
Institute of Molecular Biology and Biophysics, ETH Zürich, CH-8093 Zürich, Switzerland.